Loading necessary R packages and ERA data
This part of the document is where we actually get to the nitty-gritty of the ERA agroforestry data and therefore it requires us to load a number of R packages for both general Explorortive Data Analysis.
Loading general R packages
Show code
# Using the pacman functions to load required packages
if(!require("pacman", character.only = TRUE)){
install.packages("pacman",dependencies = T)
}
required.packages <- c("knitr","data.table", "kableExtra", #---------------------------------------------
"DT", "tidyverse", "ggExtra", # Packages needed in general
"lme4", "treemap", "pillar", "threadr",
"readr", "broom.mixed", "dotwhisker",
"skimr", "hablar", "see", "ggpubr",
"RColorBrewer", "ruler", "rstatix",
"corrr", "GGally", "cowplot", "ggridges",
"report", "rcompanion", "ggbeeswarm",
"ggfortify", "DataExplorer", "gstat",
"superheat", "patchwork", "Boruta",
"stringr", "dplyr", "see", "ggplot2",
"ggfittext", "d3treeR", "grid", "recipes")
p_load(char=required.packages, install = T,character.only = T)
Loading the ERAg package
The ERAg package includes all the ERA data we need for the EDA.
Show code
#devtools::install_github("peetmate/ERAg",
# auth_token = "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",
# build_vignettes = TRUE,
# dependencies = TRUE)
library(ERAg)
The total ERA dataset
We will first need to “split” the ERA data using the inherent ERAg function called “ERAComboSplit.” Using the ERAComboSplit function we can split practice and product combinations into duplicate individual rows each contain a unique combination of any practice x product combination present in the original observation.
ERA.Compiled.Split <- ERAComboSplit(Data = ERAg::ERA.Compiled)
# We count the number of studies per level of the practice hierarchy (see PracticeCodes
# object for more information on the practice hierarchy)
ERA.Compiled.Split.Pracs <- ERA.Compiled.Split[,list(N.Studies=length(unique(Code))),
by=list(SubPrName.Combo,PrName.Combo,Theme.Combo)]
# Visualize with the treemap function proportions of ERA Theme
# png(filename="tree.png",width=800, height=800)
ERA.tree.theme.and.practices <- (treemap::treemap(ERA.Compiled.Split.Pracs,
index=c("Theme.Combo","PrName.Combo"),
vSize="N.Studies",
type="index",
palette = "Set3",
border.col=c("black","white"),
border.lwds=c(5,1),
fontsize.title=12,
title="Proportion of ERA theme and practices
based on the number of studies for each ERA theme and practice"))
# Interactive treemap using the d3treeR package function d3tree()
ERA.tree.theme.and.practices.d3viz <- d3tree(ERA.tree.theme.and.practices,
id = "name",
celltext = "name",
valueField = "size",
width = 800, height = 600,
rootname = "ERA Themes and Practices")
Lets explore the proportions of data in ERA for each Practice under each Theme, based on the number of studies. Explorer the Treemap interactively!
Show code
Figure 1: Proportion of ERA theme and practices
Visualising each ERA theme and practice using a tree map gives a good understanding of the proportions of data under each theme and for each practice in the ERA data - based on the number of studies. We see agroforestry, our focus only accounts for a limited amount of the total ERA data.
Agroforestry data within ERA
Let us now focus on the agroforestry data within ERA by selecting data from ERA.Compiled that are only found under the ERA Theme “Agroforestry.”
af <- ERAg::ERA.Compiled[grepl("Agroforestry" , Theme)]
dim(af)
[1] 9605 144Now we can view the subsetted agroforestry data.
Show code
rmarkdown::paged_table(af)
Index <int> | Code <chr> | Author <chr> | Date <int> | Journal <chr> | DOI <chr> | |
|---|---|---|---|---|---|---|
| 2580 | NN0045 | Kho RM | 2001 | AGROFOREST SYST | 10.1023/a:;1011820412140 | |
| 2581 | NN0045 | Kho RM | 2001 | AGROFOREST SYST | 10.1023/a:;1011820412140 | |
| 2582 | NN0045 | Kho RM | 2001 | AGROFOREST SYST | 10.1023/a:;1011820412140 | |
| 2583 | NN0045 | Kho RM | 2001 | AGROFOREST SYST | 10.1023/a:;1011820412140 | |
| 2584 | NN0045 | Kho RM | 2001 | AGROFOREST SYST | 10.1023/a:;1011820412140 | |
| 2589 | NN0045 | Kho RM | 2001 | AGROFOREST SYST | 10.1023/a:;1011820412140 | |
| 2590 | NN0045 | Kho RM | 2001 | AGROFOREST SYST | 10.1023/a:;1011820412140 | |
| 2769 | NN0048 | Lamers JPA | 1995 | TROPENLANDWIRT | ||
| 2770 | NN0048 | Lamers JPA | 1995 | TROPENLANDWIRT | ||
| 2772 | NN0048 | Lamers JPA | 1995 | TROPENLANDWIRT |
Show code
af %>%
filter(Product.Simple == "Cassava") %>%
group_by(Code)
A tibble: 315 × 144
Groups: Code [18]
Index Code Author Date Journal DOI Elevation Country
1 5549 NN0116 Fagbola O 1998 BIOL FER… 10.1007…
9 55213 JS0006.1 Akonde TP 1996 AGROFORE… 10.1007…
10 55258 JS0006.1 Akonde TP 1996 AGROFORE… 10.1007…
# … with 305 more rows, and 136 more variables: # ISO.3166.1.alpha.3
The total agroforestry data is fairly large and contains 9871 observations from 270 studies with a total of 142 columns. How much of the total ERA data is under the Theme “Agroforestry?” In order to answer this we are going to divide the number of observations from the agroforestry data with the total number of ERA observations -and take the percentage.
nrow(af)/nrow(ERAg::ERA.Compiled)*100
[1] 9.069963The ERA database currently has about 9 % of observations that falls under the Theme “Agroforestry.” This is fairly little as Zomer et al. (2016) mapped the extent of trees on farms on the continent using satellite imagery and geo-datasets and found that nearly 30 % of the agricultural land in Sub-Saharan Africa had at least 10 % tree cover (in both 2000 and 2010), with nearly 40% of the population living in agricultural lands are based in areas that some way is characterised by agroforestry. In addition, remote sensing data shows that on a global level, in 2010, 43% of all agricultural lands had at least 10% tree cover and that the tree cover has increased by 2% over the previous ten years. So the question is: Does the proportion of “agroforestry” data reflect the reality of farmers and agroecosystems in Africa - or should there in reality be more research on agroforestry in Africa?
| Tree Cover (%) | 2000 | 2010 | ||||
|---|---|---|---|---|---|---|
| km2 | % of Total Agricultural Land | Population (Millions) | % of Persons Who Live in Agricultural Areas | km2 | % of Total Agricultural Land | |
| >10 | 1,089,278 | 27.5 | 67.6 | 37 | 1,137,864 | 28.7 |
| >20 | 528,602 | 13.3 | 28.2 | 16 | 582,064 | 14.7 |
| >30 | 345,302 | 8.7 | 13.0 | 7 | 353,961 | 8.9 |
| Table: Population estimates were not calculated for 2010. Source: Zomer et al. (2016) |
Splitting practice and product combinations
We have to split the practice and product combinations of the agroforestry data to visualise the proportions of the individual practices, outcomes and crops within the agroforestry data.
Show code
[1] 284There are 286 sub-practice combinations in the ERA agroforestry data. Lets have a look at these practices.
Show code
# Lets view these unique sub-practice combinations
af[,unique(SubPrName)] %>% kbl() %>%
kable_paper() %>%
scroll_box(width = "100%", height = "400px")
| x |
|---|
| Parklands |
| Inputs Urea-Parklands |
| Inputs P-Parklands |
| Inputs P-Inputs Urea-Parklands |
| Windbreak |
| Mulch (nonNfix)-Windbreak |
| AgFor Prune Mulch (Nfix)-Inputs Kraaling |
| AgFor Prune Mulch (Nfix) |
| AgFor Prune Mulch (nonNfix) |
| AgFor Alley (Nfix)-AgFor Prune (Unknown)-AgFor Prune Mulch (Nfix)-Hedge |
| AgFor Alley (nonNfix)-AgFor Prune (Unknown)-AgFor Prune Mulch (nonNfix)-Hedge |
| AgFor Alley (Nfix)-AgFor Alley (nonNfix) |
| AgFor Prune (Nfix) |
| AgFor Prune (Nfix)-Inputs N |
| AgFor Alley (Nfix)-AgFor Prune (Unknown) |
| AgFor Alley (Nfix)-AgFor Prune (Unknown)-AgrFor Prune Incorp (Nfix) |
| AgrFor Prune Incorp (Nfix) |
| AgFor Alley (Nfix) |
| AgFor Prune Incorp (nonNfix) |
| AgFor Alley (Nfix)-AgFor Prune (Unknown)-AgrFor Prune Incorp (Nfix)-Inputs N |
| AgFor Alley (Nfix)-AgFor Prune (Unknown)-AgrFor Prune Incorp (Nfix)-Inputs P |
| AgFor Alley (Nfix)-AgFor Prune (Unknown)-AgrFor Prune Incorp (Nfix)-Inputs Micro-Inputs N |
| AgFor Alley (Nfix)-AgFor Prune (Unknown)-AgrFor Prune Incorp (Nfix)-Inputs Micro-Inputs N-Inputs P |
| AgFor Alley (Nfix)-AgFor Prune (Nfix)-AgFor Prune (Unknown) |
| AgFor Prune Incorp (nonNfix)-Inputs K-Inputs P |
| AgFor Prune Incorp (nonNfix)-Inputs K-Inputs N-Inputs P |
| AgFor Alley (nonNfix)-AgFor Prune (Unknown) |
| AgFor Alley (nonNfix) |
| Silvopastoral |
| AgrFor Prune Incorp (Nfix)-Inputs Micro-Inputs N |
| AgFor Prune (Unknown)-Parklands |
| AgFor Prune (Unknown) |
| AgFor Prune Mulch (Nfix)-Inputs N-Inputs P |
| AgFor Prune Mulch (Nfix)-Inputs K-Inputs N-Inputs P |
| AgFor Alley (nonNfix)-Inputs K-Inputs N-Inputs P-Inputs Urea |
| AgFor Alley (Nfix)-AgFor Prune (Unknown)-AgFor Prune Mulch (Nfix) |
| AgFor Alley (Nfix)-Inputs Urea |
| AgFor Alley (Nfix)-NoTill |
| AgFor Prune Mulch (Nfix)-Inputs N |
| AgFor Prune Mulch (Nfix)-Inputs N-Inputs Urea |
| AgrFor Prune Incorp (Nfix)-Inputs N-pH |
| AgFor Prune Mulch (Nfix)-Inputs N-pH |
| AgFor Alley (Nfix)-AgFor Prune Mulch (Nfix) |
| AgFor Prune Mulch (Nfix)-Inputs Urea |
| Hedge |
| Grass Strips-Hedge |
| AgFor Alley (nonNfix)-AgFor Prune (noID)-AgFor Prune (Unknown) |
| AgFor Alley (Nfix)-AgFor Prune (noID)-AgFor Prune (Unknown) |
| AgFor Alley (Nfix)-AgFor Alley (nonNfix)-AgFor Prune (noID)-AgFor Prune (Unknown) |
| AgFor Prune Incorp (nonNfix)-Residue Incorp (nonNfix) |
| AgFor Prune (Unknown)-AgFor Prune Incorp (nonNfix) |
| AgFor Prune (Unknown)-AgFor Prune Incorp (nonNfix)-Inputs N |
| AgFor Prune Incorp (nonNfix)-Inputs P |
| AgFor Prune (Unknown)-AgrFor Prune (nonNfix)-Residue Incorp (Nfix) |
| AgFor Prune (Unknown)-AgrFor Prune (nonNfix)-Inputs P-Residue Incorp (Nfix) |
| AgFor Prune Incorp (nonNfix)-Inputs N |
| AgrFor Prune Incorp (Nfix)-Inputs N |
| AgrFor Prune (nonNfix)-AgrFor Prune Incorp (Nfix) |
| AgFor Prune (Nfix)-AgrFor Prune Incorp (Nfix) |
| AgFor Alley (nonNfix)-AgFor Prune (Unknown)-AgFor Prune Mulch (nonNfix) |
| AgrFor Prune Incorp (Nfix)-Inputs P |
| AgFor Prune Incorp (nonNfix)-Inputs N-Inputs P |
| AgrFor Prune Incorp (Nfix)-Inputs N-Inputs P |
| AgFor Prune Incorp (nonNfix)-Inputs K-Inputs P-Inputs Urea |
| AgFor Prune Incorp (nonNfix)-Inputs Manure |
| AgFor Alley (Nfix)-AgFor Prune (Unknown)-AgrFor Prune Incorp (Nfix)-Inputs Manure |
| AgrFor Prune Incorp (Nfix)-Inputs N-Inputs P-Inputs Urea |
| AgFor Prune Incorp (nonNfix)-Inputs N-Inputs P-Inputs Urea |
| AgrFor Prune Incorp (Nfix)-Inputs Micro |
| AgFor Prune (Unknown)-AgrFor Prune Incorp (Nfix)-Inputs K-Inputs N-Inputs P |
| AgFor Prune (Unknown)-AgrFor Prune Incorp (Nfix) |
| AgFor Prune Mulch (nonNfix)-Inputs P |
| AgFor Alley (Nfix)-AgFor Prune Mulch (Nfix)-Inputs Micro-Inputs N |
| AgFor Prune Mulch (Nfix)-Inputs Micro-Inputs N |
| AgFor Alley (Nfix)-AgFor Prune Mulch (Nfix)-Inputs N |
| AgFor Prune (Unknown)-AgFor Prune Mulch (Nfix) |
| AgFor Alley (Nfix)-AgFor Prune (Unknown)-Inputs Micro-Inputs N |
| AgFor Alley (Nfix)-AgFor Alley (nonNfix)-AgFor Prune (Unknown) |
| AgFor Alley (Nfix)-AgFor Alley (nonNfix)-AgFor Prune (Unknown)-Inputs Micro-Inputs N |
| AgFor Alley (nonNfix)-AgFor Prune (Unknown)-Inputs Micro-Inputs N |
| AgFor Alley (nonNfix)-Inputs Micro-Inputs N |
| AgFor Alley (Nfix)-Inputs Micro-Inputs N |
| AgrFor Prune Incorp (Nfix)-Inputs K-Inputs P-Inputs Urea |
| AgFor Alley (Nfix)-AgFor Prune (Unknown)-AgrFor Prune Incorp (Nfix)-Ridge & Furrow-Seed Improv |
| AgFor Alley (Nfix)-AgFor Prune (Unknown)-AgrFor Prune Incorp (Nfix)-Seed Improv |
| AgrFor Prune Incorp (Nfix)-Inputs Micro-Inputs N-Inputs P |
| AgFor Prune (Unknown)-Inputs Micro-Inputs N |
| AgFor Alley (Nfix)-AgFor Prune (Unknown)-AgFor Prune Mulch (Nfix)-Inputs P |
| AgFor Alley (Nfix)-AgFor Prune (Unknown)-AgFor Prune Mulch (Nfix)-Inputs N |
| AgFor Alley (Nfix)-AgFor Prune (Unknown)-AgFor Prune Mulch (Nfix)-Inputs N-Inputs P |
| AgFor Alley (Nfix)-Seed Improv |
| AgFor Alley (Nfix)-AgFor Prune (Unknown)-AgFor Prune Mulch (Nfix)-Inputs K-Inputs N-Inputs P |
| AgFor Alley (Nfix)-AgrFor Prune Incorp (Nfix)-Inputs K |
| AgFor Alley (Nfix)-AgrFor Prune Incorp (Nfix)-Inputs K-Inputs P |
| AgFor Alley (Nfix)-AgrFor Prune Incorp (Nfix) |
| AgFor Alley (Nfix)-AgrFor Prune Incorp (Nfix)-Inputs Micro-Inputs N |
| AgFor Prune (Nfix)-AgFor Prune (Unknown)-Inputs N |
| AgFor Prune (Nfix)-AgFor Prune (Unknown) |
| AgFor Prune Mulch (noID) |
| AgFor Prune Incorp (nonNfix)-AgrFor Prune Incorp (Nfix) |
| AgFor Alley (Nfix)-AgFor Prune (Unknown)-AgFor Prune Mulch (Nfix)-NoTill |
| AgFor Alley (nonNfix)-AgFor Prune (Unknown)-AgFor Prune Mulch (nonNfix)-Inputs K-Inputs N-Inputs P-Inputs Urea |
| AgFor Alley (Nfix)-AgFor Prune (Unknown)-AgFor Prune Mulch (Nfix)-Inputs K-Inputs N-Inputs P-Inputs Urea |
| AgFor Alley (Nfix)-AgFor Prune (Unknown)-AgrFor Prune Incorp (Nfix)-Inputs K-Inputs N-Inputs P |
| AgFor Prune Mulch (Nfix)-AgFor Prune Mulch (nonNfix)-Inputs Manure |
| AgFor Prune Incorp (nonNfix)-AgrFor Prune Incorp (Nfix)-Inputs Manure-Ridge & Furrow |
| AgFor Prune Mulch (Nfix)-AgFor Prune Mulch (nonNfix)-Inputs Manure-Inputs Urea |
| AgFor Prune Incorp (nonNfix)-AgFor Prune Mulch (Nfix)-AgFor Prune Mulch (nonNfix)-AgrFor Prune Incorp (Nfix)-Inputs Urea-Ridge & Furrow |
| AgFor Prune Incorp (nonNfix)-AgrFor Prune Incorp (Nfix)-Inputs Manure-Inputs Urea-Ridge & Furrow |
| AgFor Prune Mulch (Nfix)-AgFor Prune Mulch (nonNfix) |
| AgFor Prune Incorp (nonNfix)-AgFor Prune Mulch (Nfix)-AgFor Prune Mulch (nonNfix)-AgrFor Prune Incorp (Nfix)-Ridge & Furrow |
| AgFor Prune Incorp (nonNfix)-AgrFor Prune Incorp (Nfix)-Inputs Urea-Ridge & Furrow |
| AgFor Prune Incorp (nonNfix)-AgrFor Prune Incorp (Nfix)-Ridge & Furrow |
| AgFor Prune Mulch (Nfix)-AgFor Prune Mulch (nonNfix)-Inputs Urea |
| AgFor Prune Mulch (nonNfix)-Parklands |
| AgFor Prune Mulch (Nfix)-AgFor Prune Mulch (nonNfix)-Mulch (nonNfix) |
| AgFor Prune Mulch (nonNfix)-Mulch (nonNfix) |
| AgFor Prune (Unknown)-AgFor Prune Mulch (nonNfix)-Parklands |
| AgFor Prune (Unknown)-AgFor Prune Mulch (nonNfix) |
| AgFor Prune Mulch (nonNfix)-Inputs Compost |
| AgFor Prune Mulch (nonNfix)-Inputs K-Inputs N-Inputs P-Inputs Urea |
| AgFor Alley (nonNfix)-Inputs K-Inputs N-Inputs Urea |
| AgFor Alley (noID) |
| AgFor Prune Incorp (nonNfix)-Inputs Compost |
| AgFor Alley (Nfix)-Inputs N-Seed Improv |
| AgFor Alley (Nfix)-Inputs K-Inputs N-Inputs P |
| AgFor Alley (Nfix)-Inputs N |
| AgFor Alley (Nfix)-Inputs N-Inputs P |
| Agrosilvopastoral |
| AgFor Alley (Nfix)-AgFor Prune (Unknown)-AgrFor Prune Incorp (Nfix)-Inputs Urea |
| AgFor Prune (Unknown)-Inputs Urea |
| AgFor Multistrata |
| AgFor Alley (Nfix)-Inputs K-Inputs N |
| AgrFor Prune (nonNfix)-Inputs Biosolids-Inputs Compost-Inputs Manure |
| AgrFor Prune (nonNfix)-Inputs Biosolids |
| AgrFor Prune (nonNfix) |
| AgrFor Prune (nonNfix)-Inputs Manure |
| AgFor Alley (nonNfix)-AgFor Prune (Unknown)-AgrFor Prune (nonNfix)-Inputs K-Inputs Urea-Mulch (nonNfix) |
| AgFor Alley (nonNfix)-AgFor Prune (Unknown)-AgrFor Prune (nonNfix)-Mulch (nonNfix) |
| AgFor Prune (Unknown)-Inputs K-Inputs N-Inputs P |
| Scattered Trees |
| AgFor Prune Mulch (nonNfix)-Inputs K-Inputs N-Inputs P |
| AgrFor Prune (nonNfix)-Inputs Urea |
| AgFor Prune (Nfix)-Inputs Urea |
| AgFor Prune (Unknown)-AgFor Prune Mulch (nonNfix)-Hedge-Inputs N-NoTill |
| AgFor Prune (Unknown)-AgFor Prune Mulch (nonNfix)-Hedge-NoTill |
| AgFor Prune (Unknown)-AgFor Prune Mulch (nonNfix)-Hedge |
| AgFor Prune (Unknown)-AgFor Prune Mulch (Nfix)-Hedge-Inputs N-NoTill |
| AgFor Prune (Unknown)-AgFor Prune Mulch (Nfix)-Hedge-NoTill |
| AgFor Prune (Unknown)-AgFor Prune Mulch (Nfix)-Hedge |
| AgFor Prune (Unknown)-AgFor Prune Mulch (nonNfix)-Hedge-Inputs N |
| AgFor Prune (Unknown)-AgFor Prune Mulch (Nfix)-Hedge-Inputs N |
| AgFor Fallow (Nfix)-AgFor Prune (noID) |
| AgFor Alley (Nfix)-AgFor Prune (Unknown)-AgFor Prune Mulch (noID) |
| AgFor Prune (noID) |
| AgFor Prune (noID)-Inputs N |
| AgFor Prune (noID)-AgFor Prune (Unknown) |
| Hedge-Mulch (noID) |
| AgFor Alley (nonNfix)-AgFor Prune (Unknown)-NoTill |
| AgFor Alley (nonNfix)-AgFor Prune (Unknown)-AgFor Prune Incorp (noID) |
| AgFor Alley (nonNfix)-AgFor Prune (Unknown)-AgFor Prune Mulch (noID)-NoTill |
| AgFor Alley (nonNfix)-AgFor Prune (Unknown)-AgFor Prune Mulch (noID) |
| AgFor Alley (Nfix)-AgFor Fallow (Nfix)-AgFor Prune (Unknown)-AgFor Prune Mulch (noID) |
| AgFor Alley (Nfix)-AgFor Fallow (Nfix)-AgFor Prune Mulch (noID)-Inputs K-Inputs N |
| AgFor Alley (Nfix)-AgFor Fallow (Nfix)-AgFor Prune (Unknown)-AgFor Prune Mulch (noID)-Inputs K-Inputs N |
| AgFor Prune (Unknown)-Inputs K-Inputs N |
| AgFor Alley (Nfix)-AgFor Fallow (Nfix)-AgFor Prune Mulch (noID) |
| AgFor Fallow (Nfix)-Inputs K-Inputs N-Inputs P |
| AgFor Fallow (Nfix)-AgFor Prune (Unknown)-Inputs K-Inputs N-Inputs P |
| AgFor Alley (Nfix)-AgFor Fallow (Nfix)-Inputs K-Inputs N-Inputs P |
| AgFor Alley (Nfix)-AgFor Prune Mulch (noID) |
| AgFor Alley (Nfix)-AgFor Fallow (Nfix)-AgFor Prune (Unknown)-Inputs K-Inputs N-Inputs P |
| AgFor Prune Mulch (noID)-Mulch (noID) |
| AgFor Alley (Nfix)-AgFor Prune (Unknown)-Mulch (noID) |
| AgFor Alley (Nfix)-AgFor Prune (Unknown)-AgFor Prune Mulch (noID)-Inputs K-Inputs N-Inputs P-Mulch (noID) |
| AgFor Alley (Nfix)-AgFor Prune (Unknown)-AgFor Prune Mulch (noID)-Mulch (noID) |
| AgFor Alley (Nfix)-AgFor Prune (Unknown)-Inputs K-Inputs N-Inputs P |
| AgFor Prune Mulch (noID)-NoTill |
| AgFor Alley (Nfix)-AgFor Prune (Unknown)-AgFor Prune Incorp (noID) |
| AgFor Alley (Nfix)-AgFor Alley (nonNfix)-AgFor Prune (noID) |
| AgFor Alley (Nfix)-AgFor Prune (Unknown)-AgFor Prune Incorp (noID)-Rotation (Mixed) |
| AgFor Alley (Nfix)-Rotation (Mixed) |
| AgFor Fallow (Nfix)-AgFor Prune Incorp (noID) |
| Grass Strips-Hedge-MinTill |
| AgFor Prune Incorp (noID) |
| AgFor Alley (Nfix)-AgFor Prune (Unknown)-AgFor Prune Incorp (noID)-Intercrop (Mixed) |
| AgFor Alley (Nfix)-AgFor Prune (Unknown)-AgFor Prune Incorp (noID)-Inputs N-Inputs P-Intercrop (Mixed) |
| AgFor Alley (Nfix)-AgFor Prune (Unknown)-AgFor Prune Incorp (noID)-AgFor Prune Mulch (noID)-Intercrop (Mixed) |
| AgFor Alley (Nfix)-AgFor Prune (Unknown)-AgFor Prune Mulch (noID)-Intercrop (Mixed) |
| AgFor Alley (Nfix)-AgFor Prune (Unknown)-Planting Basins |
| AgFor Alley (noID)-AgFor Prune (Unknown) |
| AgFor Alley (nonNfix)-MinTill-Mulch (noID) |
| AgFor Prune Incorp (noID)-Green Manure (Nfix; Space) |
| AgFor Prune Incorp (noID)-Green Manure (Nfix; Space)-Inputs K-Inputs N-Inputs P |
| AgFor Alley (nonNfix)-AgFor Prune (noID)-AgFor Prune (Unknown)-Rotation (Mixed) |
| AgFor Fallow (nonNfix) |
| AgFor Alley (nonNfix)-AgFor Prune (Unknown)-AgFor Prune Mulch (noID)-Inputs K-Inputs P-Inputs Urea |
| AgFor Alley (nonNfix)-AgFor Prune (Unknown)-AgFor Prune Mulch (noID)-Rotation (Mixed) |
| AgFor Alley (nonNfix)-Intercrop (Nfix) |
| AgFor Alley (nonNfix)-Rotation (Mixed) |
| AgFor Fallow (Nfix) |
| AgFor Fallow (Nfix)-Inputs K |
| AgFor Prune Incorp (noID)-Inputs P |
| AgFor Prune Mulch (noID)-Inputs P |
| AgFor Fallow (Nfix)-Ridge & Furrow |
| AgFor Alley (Nfix)-AgFor Prune (Unknown)-AgFor Prune Mulch (noID)-Inputs K-Inputs P-Inputs Urea |
| AgFor Fallow (Nfix)-Intercrop (nonNfix) |
| AgFor Alley (Nfix)-AgFor Prune (Unknown)-AgFor Prune Incorp (noID)-Intercrop (Mixed)-Ridge & Furrow-Seed Improv |
| AgFor Alley (Nfix)-AgFor Prune (Unknown)-AgFor Prune Incorp (noID)-Intercrop (Mixed)-Seed Improv |
| AgFor Prune Incorp (noID)-Green Manure (Nfix; Space)-Inputs N |
| AgFor Alley (Nfix)-AgFor Prune (Unknown)-AgFor Prune Incorp (noID)-Inputs Manure |
| AgFor Alley (nonNfix)-AgFor Prune (Unknown)-AgFor Prune Incorp (noID)-Inputs Manure |
| AgFor Alley (Nfix)-AgFor Prune (Unknown)-AgFor Prune Incorp (noID)-Inputs Urea |
| AgFor Fallow (Nfix)-Mulch (noID)-NoTill-Rotation (Mixed) |
| AgFor Fallow (Nfix)-Rotation (Mixed) |
| AgFor Alley (Nfix)-AgFor Prune (Unknown)-AgFor Prune Incorp (noID)-Intercrop (Mixed)-pH |
| AgFor Alley (Nfix)-AgFor Prune (Unknown)-AgFor Prune Incorp (noID)-Inputs P |
| AgFor Alley (Nfix)-AgFor Prune (Unknown)-AgFor Prune Incorp (noID)-Inputs Urea-Intercrop (Mixed) |
| AgFor Alley (Nfix)-AgFor Prune Incorp (noID) |
| AgFor Fallow (Nfix)-AgFor Prune Incorp (noID)-Inputs K-Inputs N-Inputs P |
| AgFor Fallow (Nfix)-AgFor Prune Incorp (noID)-NoTill |
| AgFor Fallow (Nfix)-AgFor Prune Incorp (noID)-Inputs K-Inputs N-Inputs P-NoTill |
| AgFor Fallow (Nfix)-AgFor Prune (noID)-NoTill |
| AgFor Fallow (Nfix)-AgFor Prune (Unknown)-AgFor Prune Mulch (noID) |
| AgFor Fallow (Nfix)-AgFor Prune (Unknown)-AgFor Prune Mulch (noID)-NoTill |
| AgFor Fallow (Nfix)-NoTill |
| AgFor Fallow (Nfix)-AgFor Prune (Unknown)-AgFor Prune Incorp (noID) |
| AgFor Fallow (Nfix)-AgFor Prune (Unknown)-AgFor Prune Incorp (noID)-Imp Fallow (Nfix)-Residue Incorp (noID) |
| AgFor Prune (Unknown)-Hedge |
| AgFor Prune (Unknown)-Hedge-Inputs N-Inputs P |
| AgFor Prune (Unknown)-Inputs N-Inputs P |
| AgFor Alley (Nfix)-AgFor Prune (Unknown)-AgFor Prune Incorp (noID)-Intercrop (nonNfix) |
| AgFor Alley (Nfix)-AgFor Prune (Unknown)-AgFor Prune Incorp (noID)-Inputs K-Inputs N-Inputs P-Intercrop (nonNfix) |
| AgFor Alley (Nfix)-AgFor Prune (Unknown)-AgFor Prune Incorp (noID)-Inputs K-Inputs N-Inputs P |
| AgFor Alley (Nfix)-AgFor Prune (Unknown)-AgFor Prune Incorp (noID)-Intercrop (nonNfix)-Residue Incorp (noID) |
| AgFor Alley (Nfix)-AgFor Prune (Unknown)-AgFor Prune Incorp (noID)-Inputs K-Inputs N-Inputs P-Intercrop (nonNfix)-Residue Incorp (noID) |
| AgFor Prune (Unknown)-AgFor Prune Mulch (noID)-Hedge |
| AgFor Prune Mulch (noID)-Intercrop (nonNfix) |
| AgFor Fallow (Nfix)-AgFor Prune (Unknown)-Ridge & Furrow |
| AgFor Fallow (Nfix)-AgFor Prune (Unknown)-Inputs Manure-Ridge & Furrow |
| AgFor Prune (Unknown)-Inputs Manure |
| AgFor Fallow (Nfix)-AgFor Prune (Unknown)-Inputs K-Inputs N-Inputs P-Seed Improv |
| AgFor Fallow (Nfix)-Inputs K-Inputs N-Inputs P-Seed Improv |
| AgFor Prune (Unknown)-Intercrop (nonNfix) |
| AgFor Alley (Nfix)-Intercrop (nonNfix) |
| AgFor Alley (nonNfix)-Intercrop (nonNfix) |
| AgFor Alley (Nfix)-AgFor Prune (Unknown)-AgFor Prune Incorp (noID)-Inputs Urea-Rotation (Mixed) |
| AgFor Multistrata-Intercrop (Mixed) |
| AgFor Alley (nonNfix)-Intercrop (Mixed) |
| AgFor Multistrata-Inputs Manure-Intercrop (Mixed) |
| AgFor Alley (Nfix)-Intercrop (Mixed)-Seed Improv |
| AgFor Alley (Nfix)-Intercrop (Mixed) |
| AgFor Alley (Nfix)-Rotation (Mixed)-Seed Improv |
| AgFor Alley (Nfix)-Inputs K-Inputs N-Inputs Urea-Intercrop (Mixed) |
| AgFor Alley (Nfix)-Inputs K-Inputs N-Inputs Urea-Intercrop (Mixed)-Seed Improv |
| AgFor Alley (Nfix)-Inputs K-Inputs N-Inputs Urea-Rotation (Mixed)-Seed Improv |
| AgFor Alley (nonNfix)-AgFor Prune (noID)-AgFor Prune (Unknown)-Inputs K-Inputs Urea-Mulch (noID) |
| AgFor Alley (nonNfix)-AgFor Prune (noID)-AgFor Prune (Unknown)-Mulch (noID) |
| AgFor Alley (Nfix)-AgFor Prune (Unknown)-AgFor Prune Incorp (noID)-Inputs K-Inputs N-Inputs P-Inputs Urea |
| AgFor Alley (nonNfix)-Green Manure (nonNfix; Time)-Rotation (nonNfix) |
| AgFor Fallow (Nfix)-AgFor Fallow (nonNfix) |
| AgFor Alley (Nfix)-AgFor Fallow (Nfix)-AgFor Fallow (nonNfix)-AgFor Prune (noID)-AgFor Prune (Unknown) |
| AgFor Alley (Nfix)-AgFor Fallow (Nfix)-AgFor Fallow (nonNfix) |
| AgFor Alley (Nfix)-AgFor Fallow (Nfix)-AgFor Prune (Unknown) |
| AgFor Alley (nonNfix)-AgFor Fallow (nonNfix)-AgFor Prune (Unknown) |
| AgFor Alley (nonNfix)-AgFor Fallow (nonNfix) |
| AgFor Alley (nonNfix)-AgFor Fallow (nonNfix)-AgFor Prune (Unknown)-AgFor Prune Mulch (noID) |
| AgFor Prune (Unknown)-AgFor Prune Mulch (noID)-Hedge-Inputs N-NoTill |
| AgFor Prune (Unknown)-AgFor Prune Mulch (noID)-Hedge-NoTill |
| AgFor Prune (Unknown)-AgFor Prune Mulch (noID)-Hedge-Inputs N |
| AgFor Prune (Unknown)-AgFor Prune Mulch (noID)-Hedge-Inputs K-Inputs N-Inputs P |
| AgFor Prune (Unknown)-AgFor Prune Mulch (noID)-Hedge-Inputs K-Inputs P |
| AgFor Alley (Nfix)-AgFor Prune (Unknown)-AgFor Prune Mulch (noID)-Inputs K-Inputs N-Inputs P |
| AgFor Alley (nonNfix)-AgFor Prune (Unknown)-AgFor Prune Mulch (noID)-Inputs K-Inputs N-Inputs P |
| AgFor Prune Mulch (noID)-Imp Fallow (Nfix)-Intercrop (Mixed) |
| AgFor Prune Mulch (noID)-Imp Fallow (Nfix)-Inputs Manure-Intercrop (Mixed) |
| AgFor Prune Mulch (noID)-Imp Fallow (Nfix)-Inputs K-Inputs N-Inputs P-Inputs Urea-Intercrop (Mixed) |
| Inputs Compost-Scattered Trees |
| AgFor Other |
| AgFor Other-NoTill |
| AgFor Other-Mulch (nonNfix)-NoTill |
| AgFor Prune Mulch (Nfix)-Imp Fallow (Nfix) |
| AgFor Alley (Mixed) |
| AgFor Prune Mulch (noID)-Inputs K-Inputs P |
| AgFor Prune Mulch (noID)-Inputs K-Inputs N-Inputs P |
| AgFor Prune Mulch (nonNfix)-MinTill-Planting Basins |
| AgFor Prune Mulch (Nfix)-MinTill-Planting Basins |
| AgFor Prune Mulch (nonNfix)-NoTill |
| AgFor Prune Mulch (nonNfix)-Intercrop (Mixed)-NoTill |
| AgFor Prune Mulch (Nfix)-AgFor Prune Mulch (noID)-Inputs Urea |
| AgFor Prune Mulch (Nfix)-AgFor Prune Mulch (noID) |
| AgrFor Prune Incorp (Nfix)-Inputs Urea |
| AgFor Alley (nonNfix)-AgFor Prune Mulch (noID) |
| AgFor Prune Mulch (noID)-Intercrop (Mixed)-Mulch (nonNfix) |
| AgFor Prune Incorp (noID)-Scattered Trees |
| AgFor Prune Mulch (nonNfix)-Mulch (noID) |
| AgFor Prune (Unknown)-Inputs N |
| AgFor Alley (Nfix)-AgFor Prune Mulch (noID)-MinTill |
| AgFor Alley (Nfix)-AgFor Prune Mulch (noID)-NoTill |
| AgFor Prune Mulch (nonNfix)-Inputs P-Residue Unkn (nonNfix; NoFate) |
| AgFor Prune Mulch (nonNfix)-Inputs Manure-Residue Unkn (nonNfix; NoFate) |
| AgFor Prune (Nfix)-Green Manure (Nfix; Time) |
| AgFor Alley (Nfix)-Bunds-Trench |
| AgFor Prune Incorp (noID)-Parklands |
| AgFor Multistrata-Inputs K-Inputs N-Inputs P |
| AgFor Multistrata-Inputs K-Inputs Micro-Inputs P |
| AgFor Prune Mulch (noID)-Imp Fallow (Nfix)-Inputs K-Inputs P |
| AgFor Prune Mulch (noID)-MinTill-Mulch (noID)-NoTill |
The ERAComboSplit function inherent in ERAg package, was specially developed for splitting the ERA sub-practices. By using it on our agroforestry data we can get to know the proportion different practices and sub-practices, products and outcomes within the agroforestry data and again use the tree map function to visualise it. This is possible because ERA data is grouped or nested in a hierarchical (or tree-based) structure, we illustrate the proportions based on the number of studies for each category and sub-category and we then thow this as represented within the agroforestry data (ERA Theme: “Agroforestry”).
Lets split the practice, sub-practice combinations from the agroforestry data using the ERAComboSplit function
af.split <- ERAComboSplit(Data = af)
af.split
Agroforestry data: practices and sub-practices
Proportions of unique practices and sub-practices
Show code
af.split.subprac <- af.split[,list(N.Studies=length(unique(Code))),by=list(PrName, SubPrName)]
# Visualize with the treemap function
af.split.subprac.tree <- treemap::treemap(af.split.subprac,
index=c("PrName", "SubPrName"),
vSize="N.Studies",
type="index",
palette = "Set2",
border.col=c("black","white"),
border.lwds=c(5,1),
fontsize.title=12,
title="Proportions of unique practices and subpractices
within the agroforestry data based on number of studies")
# Interactive treemap using the d3treeR package function d3tree()
af.split.subprac.tree.d3viz <- d3tree(af.split.subprac.tree,
id = "name",
celltext = "name",
valueField = "size",
width = 600, height = 800,
rootname = "Agroforestry Practices and Sub-Practices")
Explore the interactive tree map on the proportions of unique Sub-Practices nested within their respected Practices for the agroforestry data.
Show code
Figure 2: Proportions of unique practices and subpractices within the agroforestry data based on number of studies
We see that Agroforestry Pruning and Agroforestry Pruning-Alleycropping is the two most represented agroforestry practices in the data. Together with the two second most represented practices, Alleycropping and Agroforestry Pruning-Inorganic Fertiliser, they account for about half of all the data. It shows us that most research is being conducted on these practices where other agroforestry practices, such as Parklands
Agroforestry data: sub-outcomes and practices
Proportions of Sub-Outcomes and Practices
Show code
# First we count the number of studies per level of the practice hierarchy (see PracticeCodes object for more information on the practice hierarchy)
af.split.outcome <- af.split[,list(N.Studies=length(unique(Code))),by=list(PrName, Out.SubInd)]
# Visualize with the treemap function
af.split.subout.prac.tree <- treemap::treemap(af.split.outcome,
index=c("Out.SubInd", "PrName"),
vSize="N.Studies",
type="index",
palette = "Set2",
border.col=c("black","white"),
border.lwds=c(5,1),
fontsize.title=12,
title="Proportions of unique sub-outcome and practices within the agroforestry data based on number of studies")
# Interactive treemap using the d3treeR package function d3tree()
af.split.subout.prac.tree.d3viz <- d3tree(af.split.subout.prac.tree,
id = "name",
celltext = "name",
valueField = "size",
width = 800, height = 600,
rootname = "Agroforestry Sub-Otcomes and Practices")
This treemap show the proportions of unique Sub-Outcomes nested within their respected Practices for the agroforestry data. As a lot of the data does not have specific sub-outcomes for the practice we can better view this in a static way.
Show code
# First we count the number of studies per level of the practice hierarchy (see PracticeCodes object for more information on the practice hierarchy)
af.split.outcome <- af.split[,list(N.Studies=length(unique(Code))),by=list(PrName, Out.SubInd)]
# Visualize with the treemap function
af.split.subout.prac.tree <- treemap::treemap(af.split.outcome,
index=c("Out.SubInd", "PrName"),
vSize="N.Studies",
type="index",
palette = "Set2",
border.col=c("black","white"),
border.lwds=c(5,1),
fontsize.title=12,
title="Proportions of unique sub-outcome and practices within the agroforestry data based on number of studies")
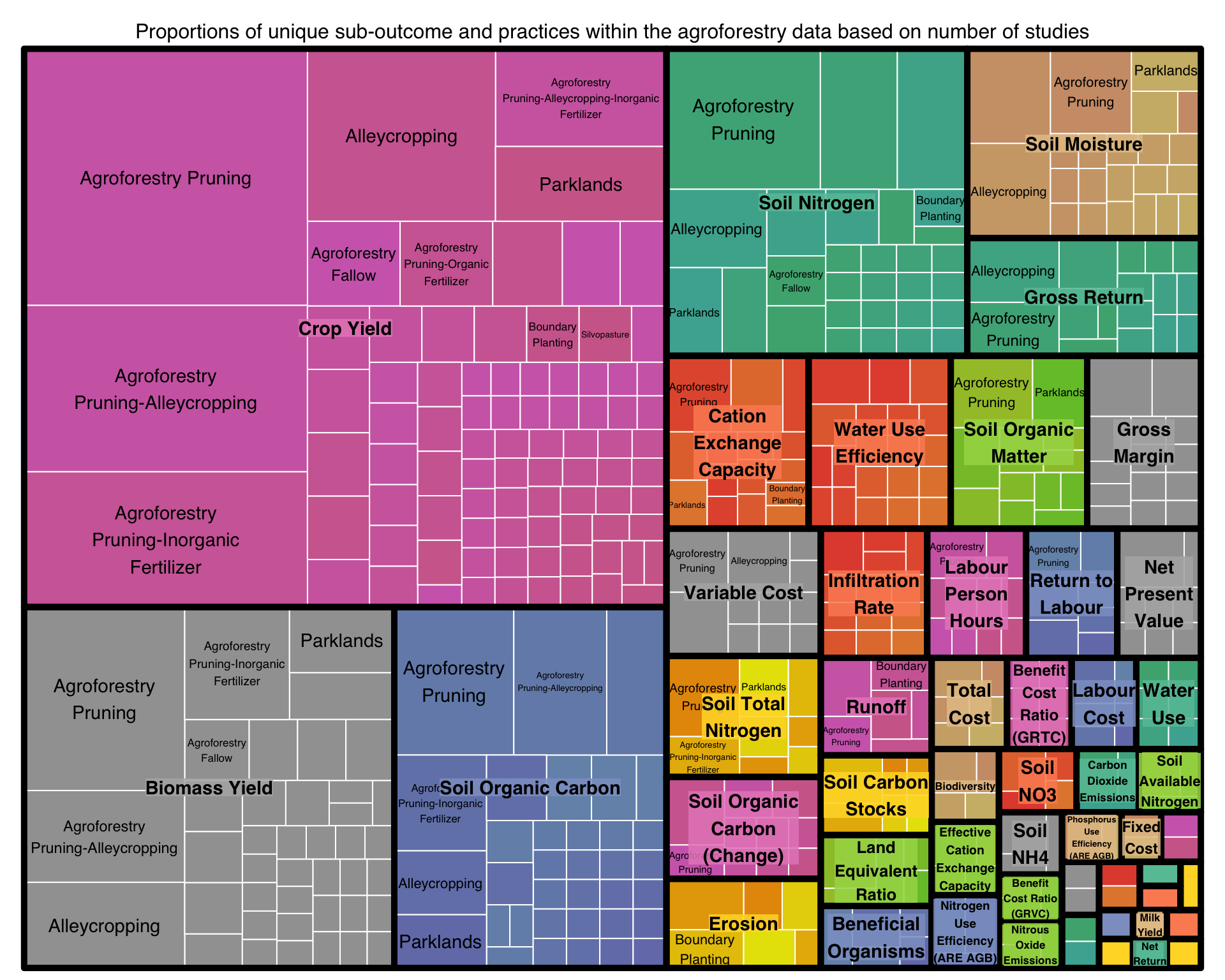
Figure 3: Proportions of unique practices and subpractices within the agroforestry data based on number of studies
Within the agroforestry data we find that the common agronomic terms crop -and biomass yields are the most represented outcomes. Hence a lot of data are pressent for these kind of outcomes. In contrast, there is little data on outcomes such as runoff or biodiversity. This indicates that any further analysis we wish to perform using machine learning techniques would benefit greatly from using data rich areas such as crop -and/or biomass yield outcomes.
Agroforestry data: Crops
Proportions of Crops represented within each Practice for the agroforestry data
This treemap show the proportions of different crops within the agroforestry data. Again its better to view this in a static way:
Show code
af.split.crops <- af.split[,list(N.Studies=length(unique(Code))),by=list(PrName, Product.Simple)]
# Visualize with the treemap function
af.split.crops.tree <- treemap::treemap(af.split.crops,
index=c("Product.Simple", "PrName"),
vSize="N.Studies",
type="index",
palette = "Set2",
border.col=c("black","white"),
border.lwds=c(5,1),
fontsize.title=12,
title = "Proportions of different crops within the agroforestry data based on number of studies")
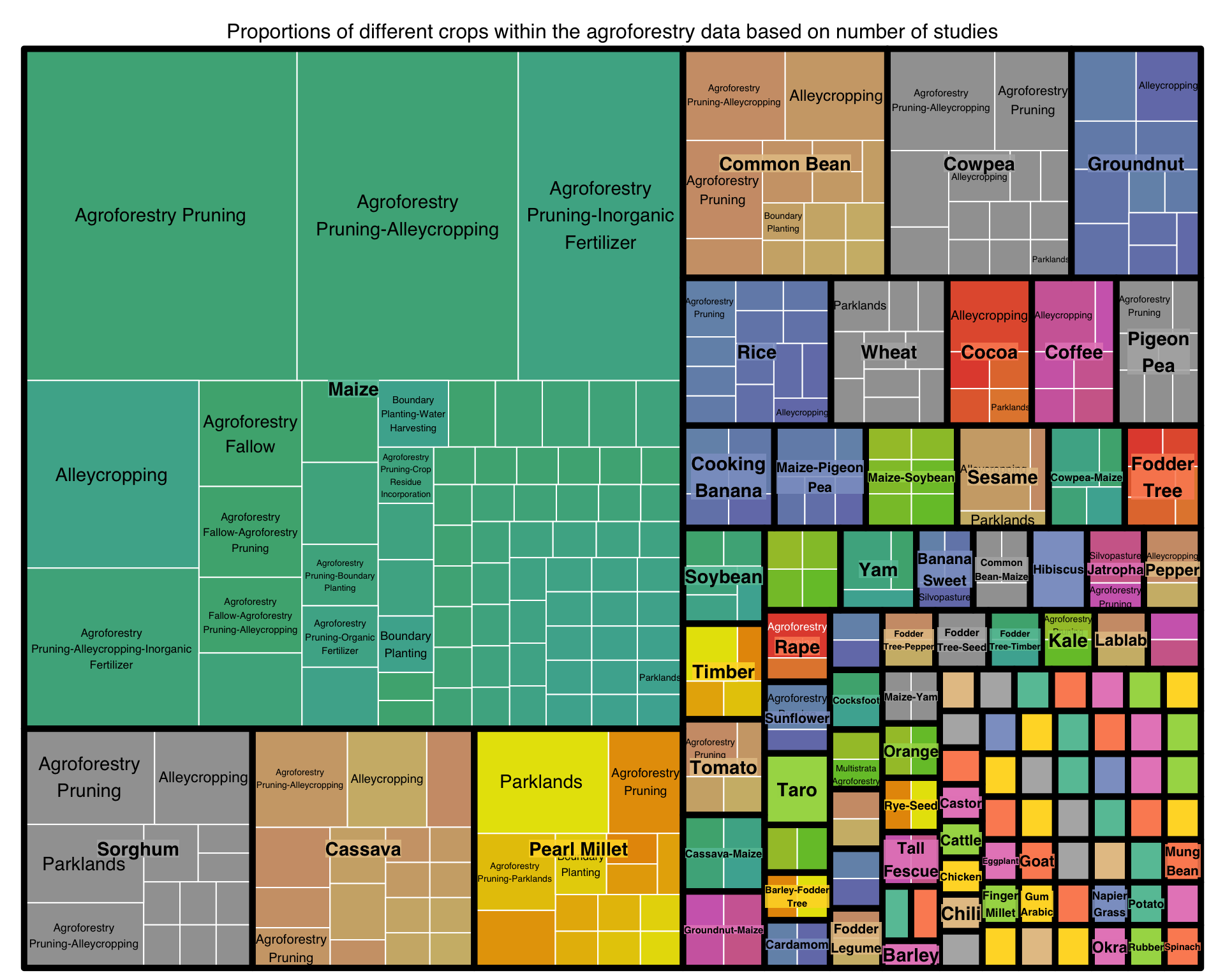
Figure 4: Proportions of Crops represented within each Practice within the agroforestry data based on number of studies
We find that the majority of crops are cereals and mostly annual crops. Maize is the most represented (the largest proportion) of all crops. This is in line with agricultural statistics from FAO, 2021 Maize is grown on over 40 M ha of land in Africa and is the primary cereal grown in over half of the countries in Africa, and one of the top two cereals in over three-quarters of these countries.
Note: Interesting to notice is that there is a great variety of crops. Many annuals, especially cereals but alo many crops we normally associate with being biannual or perennial, such as yam (Dioscorearotundata), banana (genus: Musa) and coffee (genus: Coffea).
Agroforestry data: Geographic distribution
We can use the ERAHexPlot function, inherent in the ERAg package, to spatially project where in Africa this agroforestry data comes from. We are doing this by plotting the number of studies for each ERA location on a map of the continent.
[1] 255Show code
agrofor.data.hexmap <- ERAg::ERAHexPlot(Data = af,
Showpoints = "Yes",
Point.Col = "black",
Mid = "yellow",
High = "red",)
#Warning: Ignoring unknown aesthetics: x, y
Show code
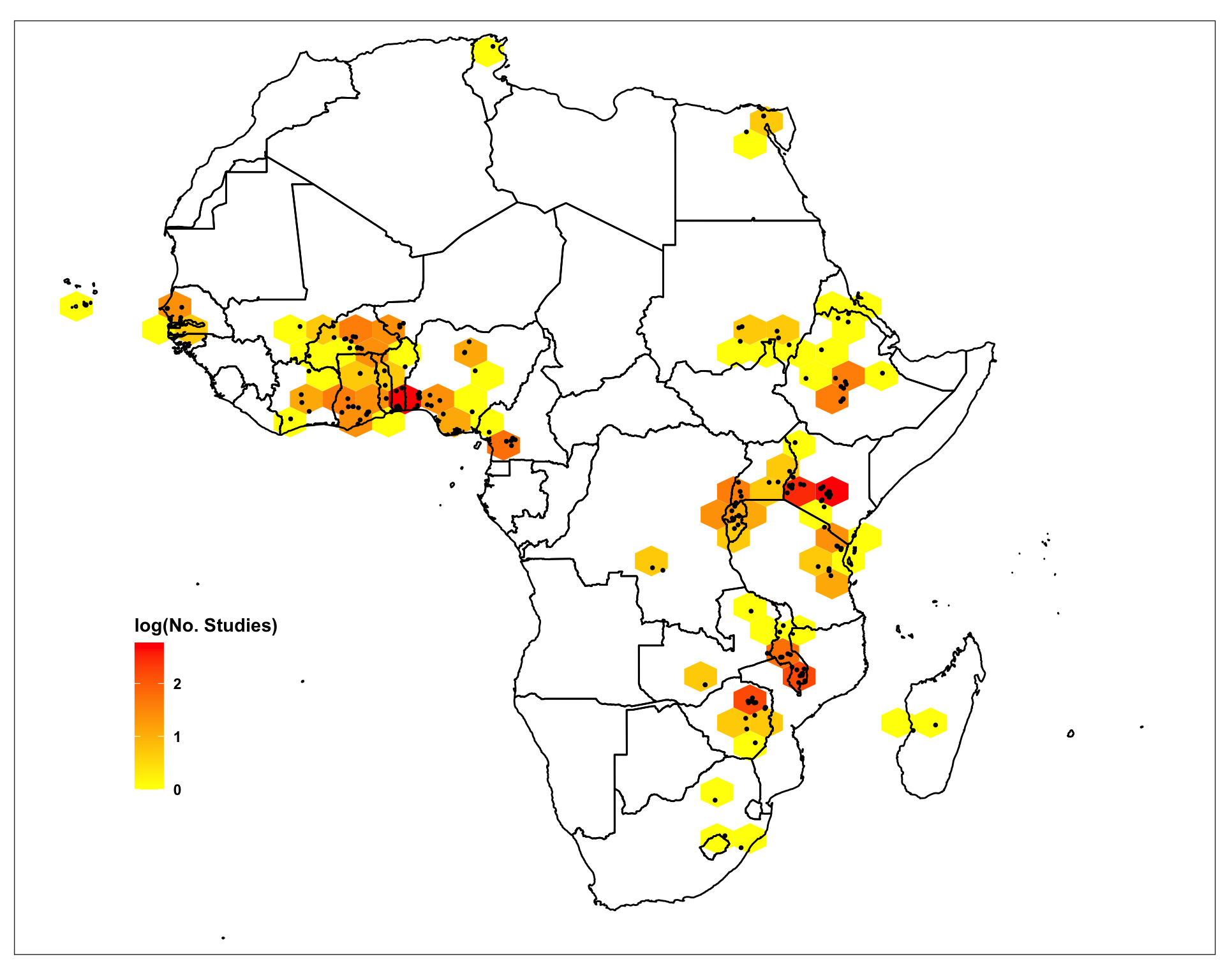
Figure 5: Spatial distribution of ERA Agroforestry studies
Countries where the agroforestry data comes from
What countries is contributing most agroforestry observations. Lets view countries and their respected proportional contribution to the ERA agroforestry data:
Show code
# (100*(af[,table(Country)] / nrow(af)) %>% round(digits=3))
af.obs.country.toplot <- af %>%
group_by(Country) %>%
summarise(count = n()) %>%
mutate(prop = count / sum(count)) %>%
arrange(desc(count))
rmarkdown::paged_table(af.obs.country.toplot)
Show code
# A tibble: 28 × 3
Country count prop
<chr> <int> <dbl>
1 Nigeria 1828 0.185
2 Zimbabwe 1603 0.162
3 Kenya 1366 0.138
4 Malawi 573 0.0580
5 Burkina Faso 523 0.0530
6 Ghana 453 0.0459
7 Benin 444 0.0450
8 Rwanda 437 0.0443
9 Cameroon 368 0.0373
10 Uganda 308 0.0312
# … with 18 more rowsWe can also plot this in a bar chart:
Show code
ggplot(af.obs.country.toplot,
(aes(x = reorder(Country, -count), y = count, fill = Country, palette = "Set2"))) +
geom_bar(stat = "identity") +
ggtitle("Number of observations per country for ERA agroforestry data") +
xlab("Number of observations") +
ylab("Country") +
theme_lucid(axis.text.angle = 45, legend.position = "none") +
theme(plot.title = element_text(size = 20, face = "bold"))
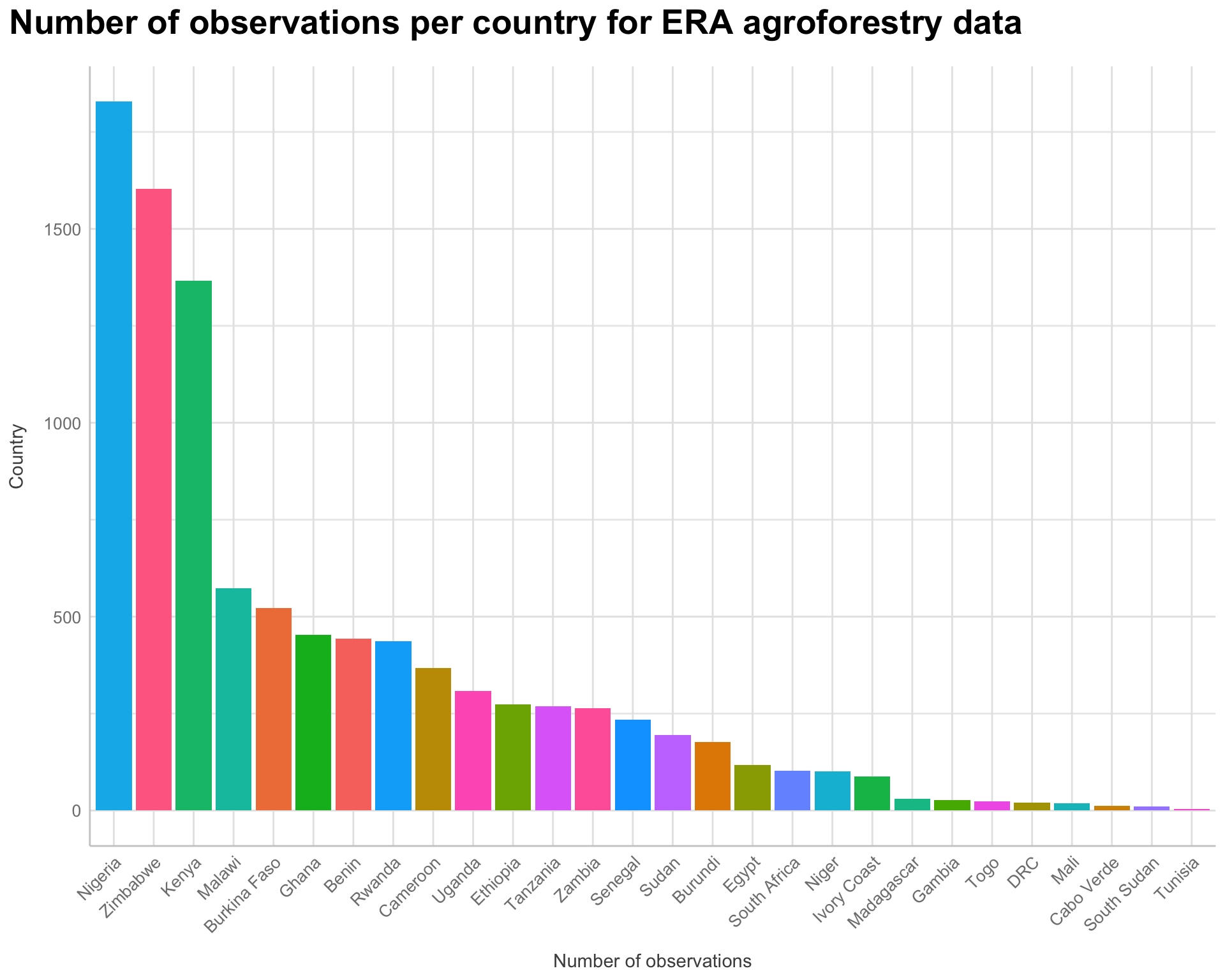
Figure 6: countries and their respected proportional contribution to the ERA agroforestry data
We see that Nigeria, Zimbabwe and Kenya are the countries where most of the ERA agroforestry data is coming from.
ERA data on crop life span
The goal is to subset the agroforestry data in a way so that is only include annual crops.
Before we proceed we will have to first subset the ERA Agroforestry data to only include annual crops. Why is that? It is simply because it is not meaningful to perform any analysis on data that have significantly different crop life spans. Just imagine if we were to compare the effect of agroforestry on the yield of cacao (Theobroma cacao) with the effect of agroforestry on the yield of potato (Solanum tuberosum). The cacao have obviously a very different growth and development cycle compared to the potato and we cannot simply aggregate the two and perform the same kind of analysis.
We are going to subset the agroforestry data, so we only proceed with data that have annual crops. We can use the information on crops life span from the ERA_EcoCrop dataset, that is part of ERA’s ERAg package. First, we are going to identify the individual experimental units (EU) of the ERA.Compiled dataset through another inherent ERA dataset, called EUCodes. The EUCodes data contains information on the unique products withing ERA. We are then creating a new dataframe with both EU codes and scientific species names (ECOCROP.Name). Finally we will use this newly created dataset to merge with the observations in the ERA.Compiled dataset.
Show code
# Identifying EU codes from the ERA.Compiled data
CODES <- ERAg::ERA.Compiled[, EU]
# Matching EU codes from the ERAg::EUCodes dataset and the EUCodes$ECOCROP.Name column
EUCodes <- data.table(ERAg::EUCodes)
ECONAME <- EUCodes[match(CODES, EU), ECOCROP.Name]
# Matching similar scientific crop species names ("ECOCROP.Name" and "species")
ERA_EcoCrop_matched <- ERAg::ERA_EcoCrop[match(ECONAME, species), Life.span]
eco <- ERA_EcoCrop %>%
dplyr::select(species, Life.span) %>%
count(species, Life.span, sort = T)
The ERA_EcoCrop dataset now contains a column with information on the scientific crop species and a column with the associated life span of that particular crop.
Show code
eco <- readRDS(here::here("ERAAnalyze_OUTPUT", "eco.RDS"))
rmarkdown::paged_table(eco)
species <chr> | Life.span <chr> | n <int> | ||
|---|---|---|---|---|
| Abelmoschus manihot | annual, perennial | 2 | ||
| Abelmoschus esculentus | annual | 1 | ||
| Abelmoschus moschatus | annual, biennial, perennial | 1 | ||
| Abies amabilis | perennial | 1 | ||
| Abies balsamea | perennial | 1 | ||
| Abies concolor | perennial | 1 | ||
| Abies pindrow | perennial | 1 | ||
| Abroma augustum | perennial | 1 | ||
| Abutilon theophrasti | annual | 1 | ||
| Acacia abyssinica | perennial | 1 |
We can now transfer the Life.span information from the ERA_EcoCrop data to the EUCodes by merging the two datasets on their common row names for the scientific crop species names. Next, we are going to merge the data on crop life span with the associated EU codes so that we eventually have a dataset that we are able to merge with the ERA.Compiled dataset based on EU codes.
Show code
No we can finally merge our information on life span of a particular crop species with the ERA.Compiled dataset. The result is a added column to the ERA.Compiled data in which we have information on whether a particular crop is annual and/or biannual and/or perennial. We merge by EU codes.
Show code
ERA_Compiled_LifeSpan <- left_join(ERA.Compiled, EcoCrop_EU, by = "EU")
era.compiled.life.span <- ERA_Compiled_LifeSpan %>%
dplyr::relocate(Index, Code, Country, EU, PrName.Code, SubPrName.Code,
ECOCROP.Name, Life.span)
During the use of left_join() function columns with similar names were automatically added a suffix of either “.x” or “.y” in order to make them unique. Before we can proceed to analyse the data we need to clean the ERA_Compiled_LifeSpan data by removing the redundant columns that were created.
Show code
# Un-comment to view the redundant columns
# era.compiled.life.span.redundance <- era.compiled.life.span %>% dplyr::relocate(ends_with(".x") | ends_with(".y"))
# Removing spaces and redundant columns
ERA_Compiled_LifeSpan <- ERA_Compiled_LifeSpan %>%
rename_at(vars(ends_with(".x")),
~str_replace(., "\\..$","")) %>%
select_at(vars(-ends_with(".y")))
# combining all NA into one group, instead of two NA categories
ERA_Compiled_LifeSpan <- ERA_Compiled_LifeSpan %>%
replace_na(list(Life.span = "NA", Life.span = "NA"))
# Number of observations for each Life.span
ERA_Compiled_LifeSpan %>%
count(Life.span, sort = TRUE)
Now we have our Compiled ERA data with clean information on whether the crop is annual or not. Lets see how the life span of crops in the ERA data is distributed.
Show code
ERA_Compiled_LifeSpan.count <- readRDS(here::here("ERAAnalyze_OUTPUT", "ERA_Compiled_LifeSpan.count.RDS"))
rmarkdown::paged_table(ERA_Compiled_LifeSpan.count)
Life.span <chr> | n <int> | |||
|---|---|---|---|---|
| annual | 71845 | |||
| NA | 28074 | |||
| perennial | 6541 | |||
| annual, biennial | 967 | |||
| biennial | 243 | |||
| biennial, perennial | 207 | |||
| annual, perennial | 75 |
Visualising the proportions of represented crop life span classes in ERA
Using a visualisation, such as a bar plots we can get a good idea of the proportions of crop life span classes in ERA’s data.
Show code
# First, we will sort the Life.span column to have the number of observations in ascending order
sorted_acending <- within(ERA_Compiled_LifeSpan,
Life.span <- factor(Life.span, levels=names(sort(table(Life.span), decreasing = FALSE))))
# Counts (or sums of Life.span). Notice that we plot the bar plot with a logarithmic x-axis. That's because there are disproportionately many observations of annual compared to the other groups.
ERA_crops_LifeSpan.plot <- ggplot(sorted_acending, aes(y = Life.span, fill = Life.span)) +
scale_x_log10(
breaks = scales::trans_breaks("log10", function(x) 10^x),
labels = scales::trans_format("log10", scales::math_format(10^.x))) +
# # Number of observations in each Life.span category:
geom_bar(color="black") +
annotation_logticks(sides = "b",
short = unit(.2,"mm"),
mid = unit(1,"mm"),
long = unit(2,"mm")) +
scale_fill_brewer(palette="Set2") +
ggtitle("Distribution of observations of each Life span class in the ERA Compiled data")+
theme_lucid(legend.position="top") +
theme(legend.position = "none") +
ylab("EcoCrop life span") +
xlab("Number of observarions (log-scale)")
Bar plot of the distribution of observations within each crop life span group
Show code
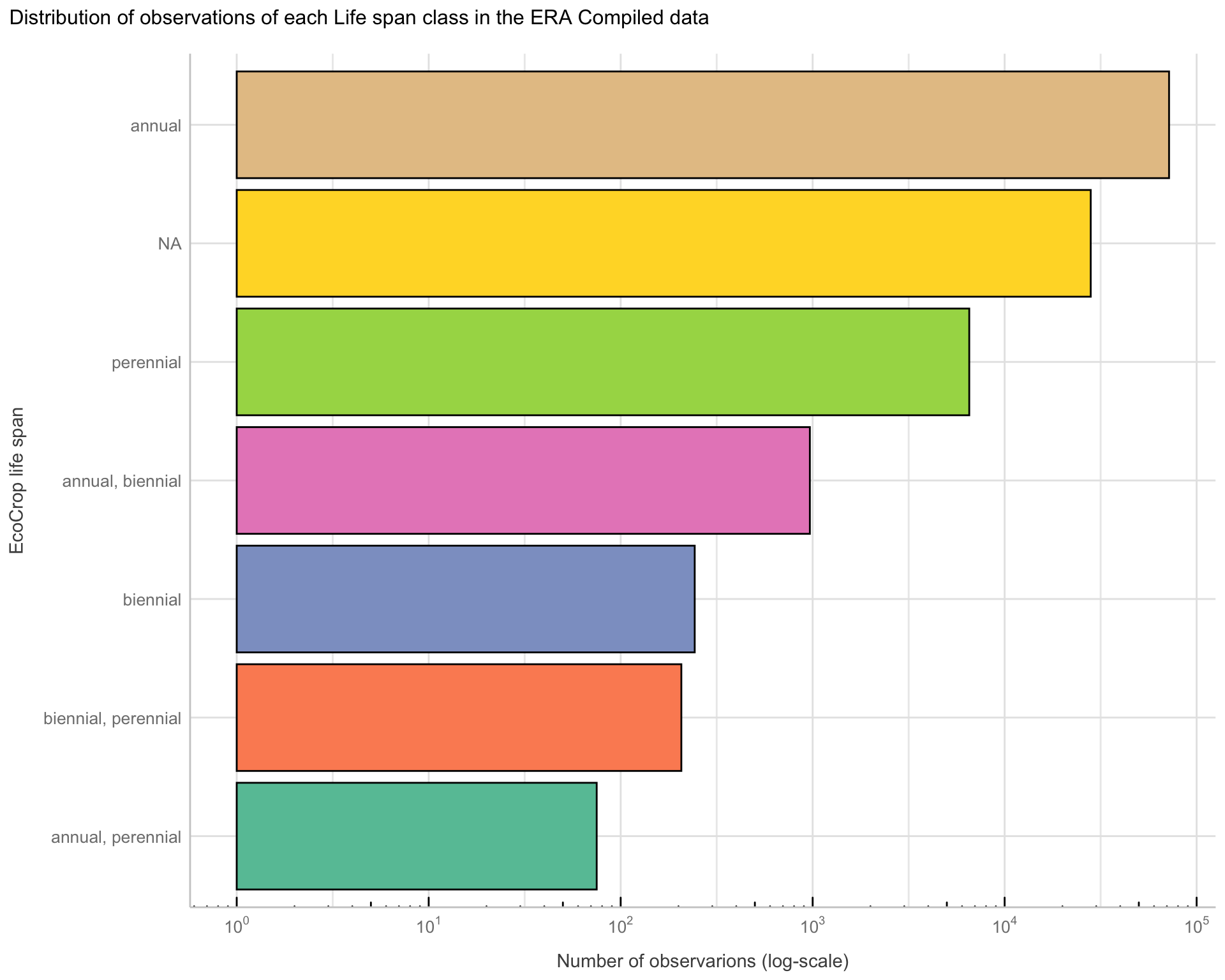
Figure 7: Distribution of Life span classes in ERA Compiled
It is evident that we have a majority of “annual” crops in ERA. However, we also see that a considerable amount of the ERA data actually lack information on whether a crop is annual or not.
Agroforestry data with only annual crops
Now that we have information on the life span of the crops in ERA we are going again select data from the ERA Theme “Agroforestry” but this time were also filtering the data on the newly added life span column. In this way we are sure that our data only include agroforestry and that this data only include annual crop species.
Show code
# Creating new agroforestry dataset
era.agroforestry <- ERA_Compiled_LifeSpan[grepl("Agroforestry", Theme)]
# Within our subsetted agroforestry dataset we are now filtering, so that we only look at annual crops.
era.agroforestry.annual <- era.agroforestry %>% filter(Life.span == "annual")
dim(era.agroforestry)
# [1] 9871 216
dim(era.agroforestry.annual)
# [1] 6745 216
| Filtering for crops = annual | Total number of observations in the dataset | Total number of column features in the dataset |
|---|---|---|
| Agroforestry data before filtering | 9871 | 216 |
| Agroforestry data after filtering | 6745 | 216 |
Show code
# Calculating the percentage decrease in observations after filtering
threadr::percentage_difference(nrow(era.agroforestry), nrow(era.agroforestry.annual))
# [1] 37.62638
By filtering out all non-annual crops we get a reduction in observations of about 37 percent. Before we proceed, lets just check that we really only have agroforestry data with annual crops.
# Checking for NA values in the data
data.table(era.agroforestry.annual)[is.na(Life.span),table(Product.Simple)]
# < table of extent 0 > <- looks good
# Checking that we only have "annual" crops
data.table(era.agroforestry.annual)[,table(Life.span)]
# Life.span
# annual
# 6745 <- looks good
The line “< table of extent 0 >” indicate that we indeed do not have any NA observations in the data. The next lines “Life.span: annual” indicate that we successfully created a data set of agroforestry data with only annual crops based on the Life.span columns.
Next, we are going to proceed with a proper analysis of the ERA agroforestry dataset using the ERAAnalyze function. This function is part of the ERAg package and serves to analyse the response ratios (RR)
It is recommended to apply some data cleaning or data preparation to the ERA dataset before feeding it into the ERAAnalyze function. Luckily there is another handy function for this crusial step in the arsenal of functions from the ERAg package. This function is funny enough called PrepareERA So before we can start using the ERAAnalyze, lets make some preparations using the PrepareERA function.
Note: Response ratios (RR) is the chosen effect size
Using the ERAAnalyze function
The ERAAnalyze function is a (meta)-analysis function that analyses outcome ratios in the ERA dataset for each combination of grouping variables as specified by column names in the Aggregate.By parameter. It is recommended to apply the ERA.Prepare function to the data before using ERAAnalyze. In detail the ERAAnalyze function performs the following to the data:
- Weighting: Individual observations are up-weighted by replication and down-weighted by the number of observations submitted from the same study, for each combination of grouping variables. Studies with more replications are likely to produce less variable information than studies with fewer. Controlling for the number of observations contributed by a study to the dataset weights each study equally. As such, outcome ratios are weighted according to the equation below, where Rep is the number of replications for RepC the control and RepE the experimental treatment, and Ns is the total number of observations contributed to the overall dataset by the study to which the observation belongs:
Weighting=((RepsE∗RepsC)/(RepsE)+(RepsC))/(Ns)
Outlier Removal: Outliers are defined using an extreme outliers method where response ratios above or below 3∗IQR (interquartile range) are removed. The ERA outcome variables analysed by this function are ratios between an experimental treatment and control outcome and should be approximately normally distributed. When the control approaches zero (e.g. yield collapse) this skews the distribution of the outcome ratio producing extremely high values tending to infinity and requiring outlier removal.
Test of Normality: A Shapiro-Wilk test is applied to raw and log-transformed outcome ratios for each combination of grouping variables. This can be used to judge whether values based on mean proportional change, mean response ratio or median proportional change should be used to evaluate practice performance.
Basic Statistical Tests: When Fast = FALSE where minimum data requirements are met linear-mixed effects or linear model is applied to the data to generate means, standard errors and variance. Linear mixed effects models use lmer::lme4 where outcomes from a grouping variable combination are from at least three sites of which two must have at least three observations. The model is weighted and includes a random intercept for site
lmer(Value≈1+(1|Site),weights=Weights))
- Correction of Jensen’s Inequality: The log-scale response ratios are back-transformed and converted to % change with and without a correction for the Jensen’s inequality. The correction applied follows the methodology from Tanadini and Mehrabi (2017) link. The use of these corrected values are recommended.
Define focus practices
What practices do we find in the ERA agroforestry data? Wee need all these practices for our ERAAnalyze function, as we are interested in the most aggregated form of agroforestry data, with all the practices!
Show code
ERAg::PracticeCodes[Theme == "Agroforestry", unique(Practice)]
[1] "FMNR" "Alleycropping"
[3] "Parklands" "Boundary Planting"
[5] "Silvopasture" "Multistrata Agroforestry"
[7] "Agroforestry Pruning" "Agroforestry Fallow"
[9] "Other Agroforestry" We define the focus practices as all nine ERA agroforestry practices and then we are going to view the number of observations for each agroforestry practice.
# Defining focus practices as all agroforestry practices
focus.prac <- c("FMNR",
"Alleycropping",
"Parklands",
"Boundary Planting",
"Silvopasture",
"Multistrata Agroforestry",
"Agroforestry Pruning",
"Agroforestry Fallow",
"Other Agroforestry")
ERAg::PracticeCodes[Practice %in% focus.prac, Definition]
# The analysis is going to include all of these nine agroforestry practices as focus practices
# Making a new dataframe with ERA agroforestry that only has annual crops
ERA.AF <- era.agroforestry.annual
# These nine practices results in the following PrName practices.
# We can run them through a add_count() function from dplyr and sort them in a
# decreasing order based on the number of observations within each PrName group.
agroforestry.prac.prname.obs <- ERA.AF %>%
dplyr::add_count(PrName, name = "n_obs_PrName")
agroforestry.prac.sort <- agroforestry.prac.prname.obs %>%
dplyr::count(PrName, n_obs_PrName, sort = TRUE)
Lets have a look.
Show code
agroforestry.prac.sort <- readRDS(here::here("ERAAnalyze_OUTPUT", "agroforestry.prac.sort.RDS"))
rmarkdown::paged_table(agroforestry.prac.sort)
PrName <chr> | |
|---|---|
| Agroforestry Pruning | |
| Agroforestry Pruning-Alleycropping | |
| Agroforestry Pruning-Inorganic Fertilizer | |
| Alleycropping | |
| Agroforestry Pruning-Alleycropping-Inorganic Fertilizer | |
| Agroforestry Fallow-Agroforestry Pruning | |
| Parklands | |
| Agroforestry Pruning-Reduced Tillage-Water Harvesting | |
| Agroforestry Fallow | |
| Agroforestry Pruning-Boundary Planting |
We see that the vast majority of observations are for the practices “Agroforestry Pruning,” “Agroforestry Pruning-Alleycropping” and “Agroforestry Pruning-Inorganic Fertilizer”
The definitions of ERA practices can be found using the function ERAg::PracticeCodes(). Here we apply the function to look at the definitions of “FMNR,” “Alleycropping” and “Multistrata Agroforestry.” The reason why we have four definitions for Alleycropping is because there are four different Sub-Practices within Alleycropping.
Show code
# Defining focus practices as all agroforestry practices
focus.prac.FNMR <- c("FMNR")
focus.prac.Alleycropping <- c("Alleycropping")
focus.prac.MultistrataAgroforestry <- c("Multistrata Agroforestry")
# Definitions of FNMR, Alleycropping and Multistrata Agroforestry
ERAg::PracticeCodes[Practice %in% focus.prac.FNMR, Definition]
ERAg::PracticeCodes[Practice %in% focus.prac.Alleycropping, Definition]
ERAg::PracticeCodes[Practice %in% focus.prac.MultistrataAgroforestry, Definition]
| Practice | Definition(s) |
|---|---|
| FMNR | “Systematic regrowth of trees or shrubs on agricultural, forest or pasture land. FMNR is used in areas where there are stumps that can coppice or seeds that can germinate. Sometimes called farmer assisted regeneration.” |
| Alleycropping | “Intercropping with rows or alleys of nitrogen fixing trees or woody shrubs.” |
| “Intercropping with rows or alleys of trees or woody shrubs that do not fix nitrogen.” | |
| “Intercropping with rows or alleys of a mixture of trees or woody shrubs of which some but not all fix nitrogen.” | |
| “Intercropping with rows or alleys of trees or woody shrubs where no information about the type of plant is given.” | |
| Multistrata Agroforestry | “Multistorey systems agroforestry systems have several spatial strata occupied by different tree (i.e. woody) crops (coffee, tea, cacao, banana etc).” |
Threshold: Minimum number of observations
Clicking the arrow to the right in the interactive table above shows us that some practices have very few numbers of observations and this can negatively impact the power and robustness of our analysis with ERAAnalyze. So let’s subset the dataset with data that have a minimum of 25 observations per practice (PrName). This is a rather arbitrary number that depends a lot on what one wish to analyse and how much data is available at first. Here we will set the threshold as a minimum number of observations to be 25.
Show code
# Loading the ERA.AF data
ERA.AF <- readRDS(here::here("ERAAnalyze_OUTPUT", "ERA.AF.RDS"))
agroforestry.prac.thresh <- ERA.AF %>%
dplyr::add_count(PrName, name = "n_obs_PrName") %>%
# only selecting PrName groups that have at least 25 observations
dplyr::filter(n_obs_PrName >= 25)
# The agroforestry data gets reduced
# dim(agroforestry.prac.prname.obs) # <- before PrName obs >= 25 threshold
# [1] 6745 217
# dim(agroforestry.prac.thresh) # <- after PrName obs >= 25 threshold
# [1] 6361 217
| Threshold | Total number of observations in the dataset | Total number of column features in the dataset |
|---|---|---|
| Before PrName obs >= 25 threshold | 6745 | 217 |
| After PrName obs >= 25 threshold | 6361 | 217 |
The threshold reduces the total dataset available for the ERAAnalyze from 6745 to 6361 observations. We did loose a bit of data but hopefully our later analysis will be more powerful and robust. This is a fine-tuning trade-off issue and one has to decide on such a threshold based on the goal of the analysis, the available data to answer the relevant questions and expert knowledge.
Note: Later we will include a second threshold to our data so that the number of studies should be of a minimum of 2 (see section Calculating response ratios with ERAAnalyze
CREATING THE NICE RIDGE LINE PLOTS OF EACH PRACTICE
Show code
ml.data.no.outliers.wf <- readRDS(here::here("ERAAnalyze_OUTPUT", "ml.data.no.outliers.wf.RDS"))
ml.data.no.outliers.wf %>%
rationalize() %>%
step_impute_mean(all_numeric()) %>%
relocate(logRR) %>%
dplyr::select(-c(Out.SubInd, Product, Site.Type, Tree, Out.SubInd.Code, Latitude, Latitude)) %>%
group_by(PrName) %>%
summarise(across(everything(), mean)) %>%
dplyr::filter(PrName == "Agroforestry Fallow" |
PrName == "Agroforestry Pruning"|
PrName == "Alleycropping"|
PrName == "Parklands") %>%
arrange(desc(logRR))
# A tibble: 4 × 39
PrName logRR Longitude Bio01_MT_Annu Bio02_MDR Bio03_Iso Bio04_TS
<fct> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 Agrofor… 0.649 9.19 18.1 11.3 56.2 326.
2 Agrofor… 0.406 19.6 NA NA NA NA
3 Parklan… -0.123 8.68 23.9 11.0 68.2 180.
4 Alleycr… -0.199 26.6 21.0 11.2 68.1 186.
# … with 32 more variables: Bio05_TWM <dbl>, Bio06_MinTCM <dbl>,
# Bio07_TAR <dbl>, Bio08_MT_WetQ <dbl>, Bio09_MT_DryQ <dbl>,
# Bio10_MT_WarQ <dbl>, Bio11_MT_ColQ <dbl>, Bio12_Pecip_Annu <dbl>,
# Bio13_Precip_WetM <dbl>, Bio14_Precip_DryM <dbl>,
# Bio15_Precip_S <dbl>, Bio16_Precip_WetQ <dbl>,
# Bio17_Precip_DryQ <dbl>, iSDA_Depth_to_bedrock <dbl>,
# iSDA_SAND_conc <dbl>, iSDA_CLAY_conc <dbl>, …Show code
ml.data.no.outliers.wf %>%
rationalize() %>%
step_impute_mean(all_numeric()) %>%
relocate(logRR) %>%
dplyr::select(-c(Out.SubInd, Product, Site.Type, Tree, Out.SubInd.Code, Latitude, Latitude)) %>%
group_by(PrName) %>%
# summarise(across(everything(), mean)) %>%
dplyr::filter(PrName == "Agroforestry Fallow" |
PrName == "Agroforestry Pruning"|
PrName == "Alleycropping"|
PrName == "Parklands") %>%
ggplot() +
geom_boxplot(aes(x = reorder(PrName, iSDA_SAND_conc), y = iSDA_SAND_conc, col = PrName)) +
geom_jitter(aes(x = reorder(PrName, iSDA_SAND_conc), y = iSDA_SAND_conc, col = PrName), height = 0.4, width = 0.1) +
theme_lucid(plot.title.size = 25,
axis.text.size = 15,
legend.title.size = 20,
legend.text.size = 20,
axis.title.size = 20) +
xlab("Agroforestry Practices") +
ylab("Sand content") +
theme(legend.position = "none", axis.text.x = element_text(angle = 60, hjust=1), plot.margin=unit(c(1,6,1,6),"cm"))
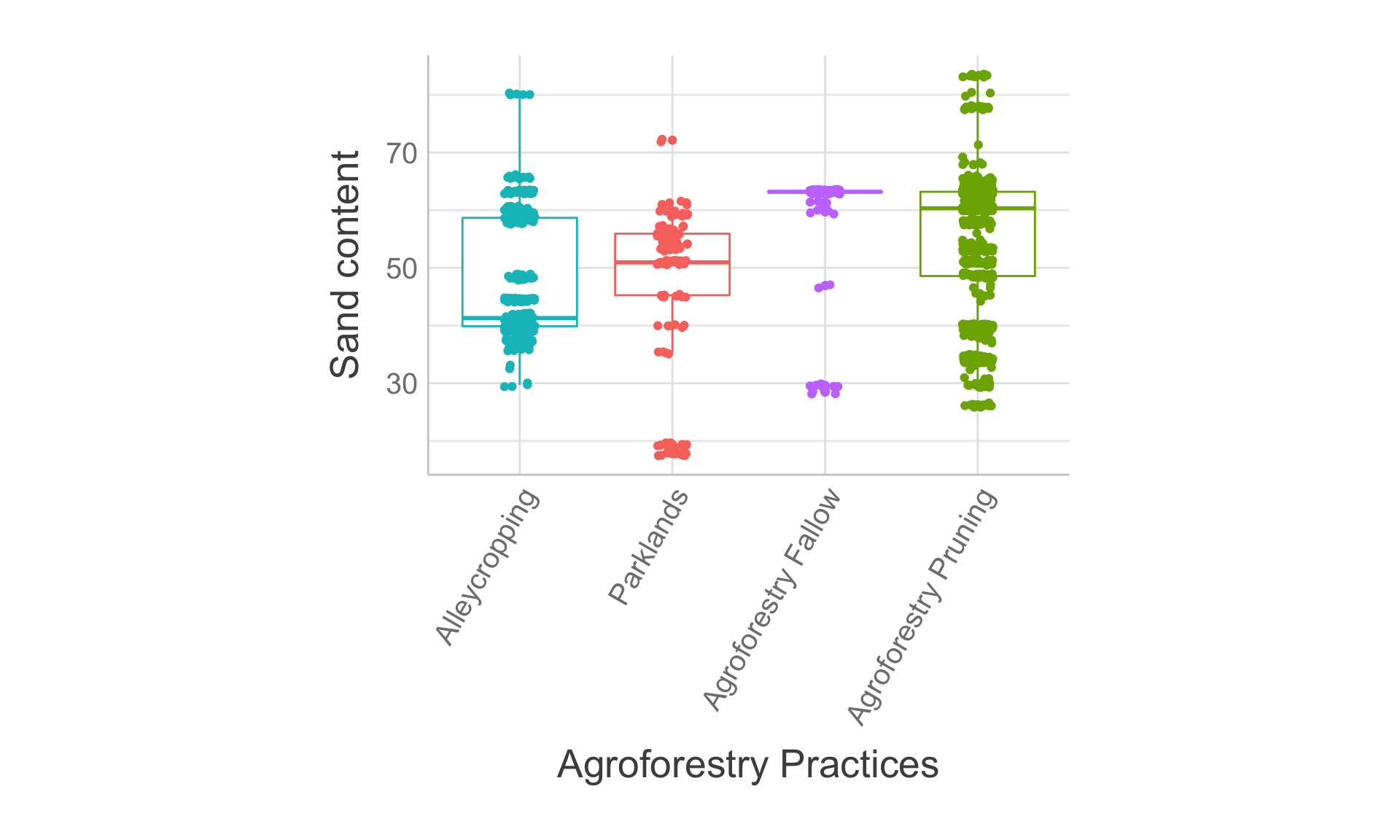
Figure 8: Agroforestry practices vs sand content
Show code
agfor_ridgeline_data <-
ml.data.no.outliers.wf %>%
rationalize() %>%
drop_na() %>%
relocate(logRR) %>%
dplyr::select(-c(Out.SubInd, Product, Site.Type, Tree, Out.SubInd.Code, Latitude, Longitude)) %>%
group_by(PrName) %>%
# summarise(across(everything(), mean)) %>%
dplyr::filter(PrName == "Agroforestry Fallow" |
PrName == "Agroforestry Pruning"|
PrName == "Alleycropping"|
PrName == "Parklands")
Show code
agfor_ridgeline_data %>%
ggplot(aes(y = PrName, x = iSDA_SAND_conc)) +
stat_density_ridges(quantile_lines = TRUE, quantiles = c(0.025, 0.50, 0.975), col = "orange", alpha = 0.5, size = 0.2, scale = 2,
jittered_points = TRUE, position = position_points_jitter(width = 5, height = 0.05),
point_shape = '|', bandwidth = 6) +
theme_lucid() +
xlab("Sand concentration %") +
ylab("") +
ggtitle("SAND concentration (%)")
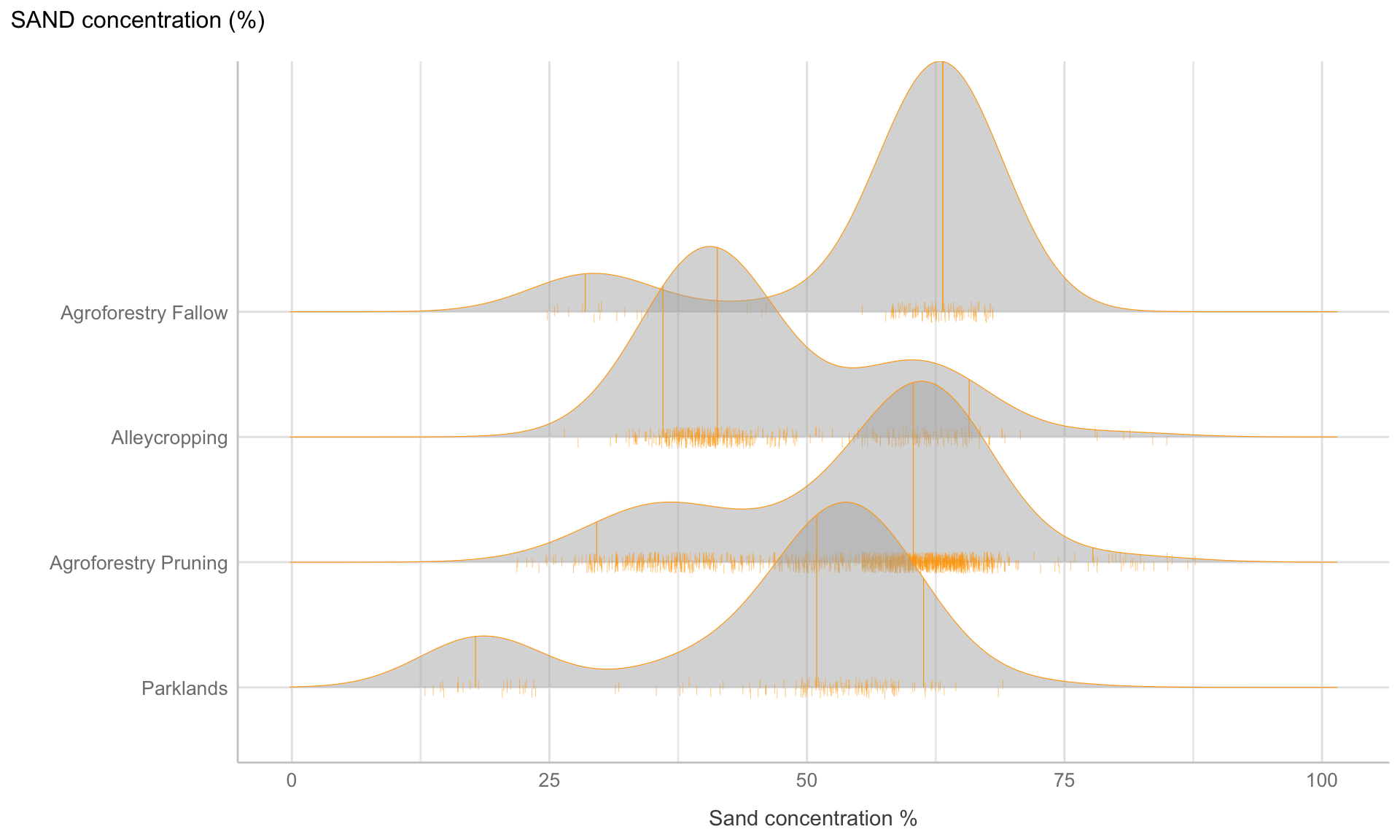
Figure 9: Ridge line plot: Distribution of sand content for the four different agroforestry practicest
Show code
agfor_ridgeline_data %>%
ggplot(aes(y = PrName, x = Bio14_Precip_DryM)) +
stat_density_ridges(quantile_lines = TRUE, quantiles = c(0.025, 0.50, 0.975), col = "midnightblue", alpha = 0.5, size = 0.2, scale = 2,
jittered_points = TRUE, position = position_points_jitter(width = 50, height = 0.05), point_shape = '|',
bandwidth = 3.75) +
theme_lucid() +
xlab("Precipitation (driest month)") +
ylab("") +
ggtitle("Precipitation (driest month) [mm]")
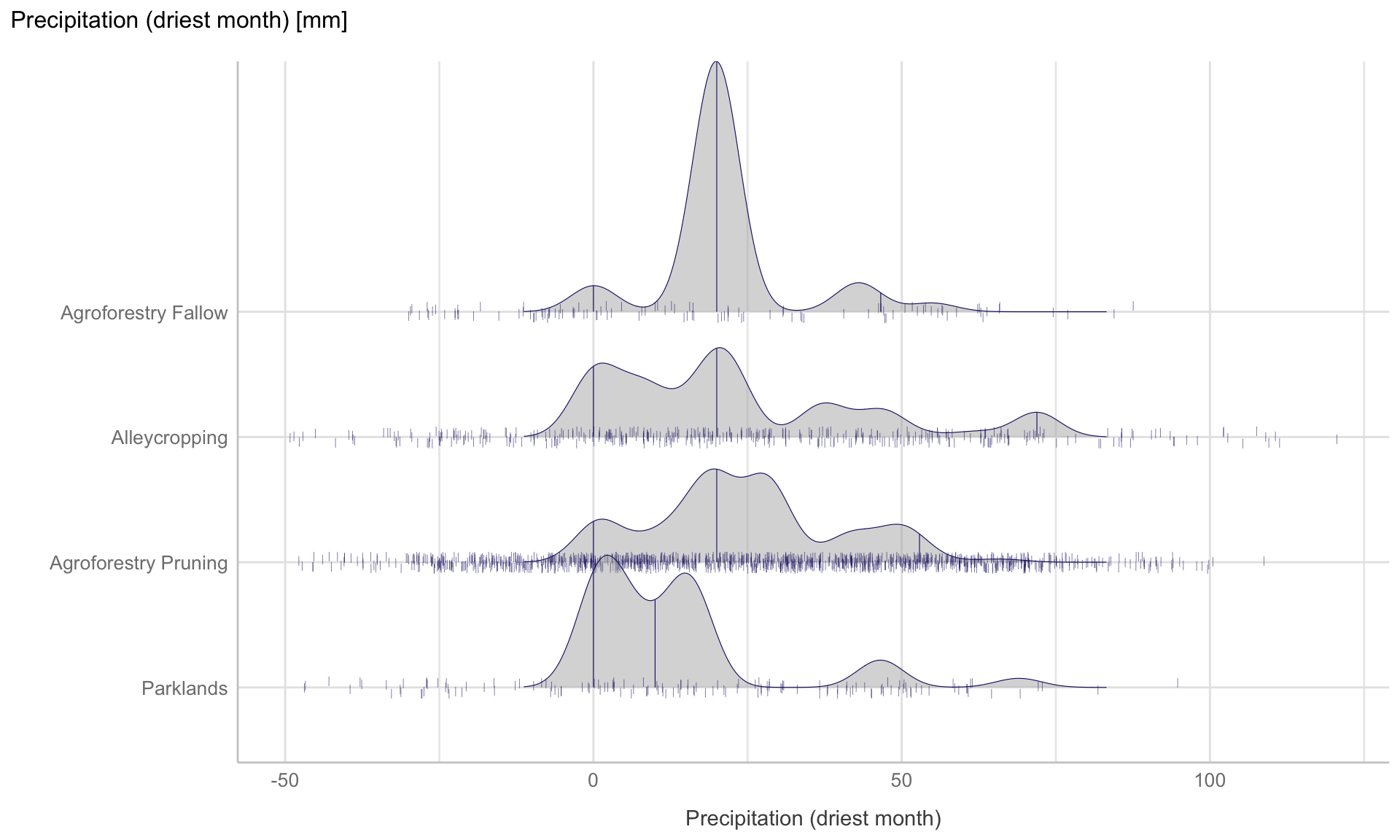
Figure 10: Ridge line plot: Distribution of precipitation of driest month for the four different agroforestry practices
Show code
agfor_ridgeline_data %>%
ggplot(aes(y = PrName, x = Bio05_TWM)) +
stat_density_ridges(quantile_lines = TRUE, quantiles = c(0.025, 0.50, 0.975), col = "darkgreen", alpha = 0.5, size = 0.2, scale = 2,
jittered_points = TRUE, position = position_points_jitter(width = 50, height = 0.05),
point_shape = '|', bandwidth = 2) +
theme_lucid() +
xlab("Temperature (max temperature of warmest month)") +
ylab("") +
ggtitle("Temperature (max temperature of warmest month) [°C]")
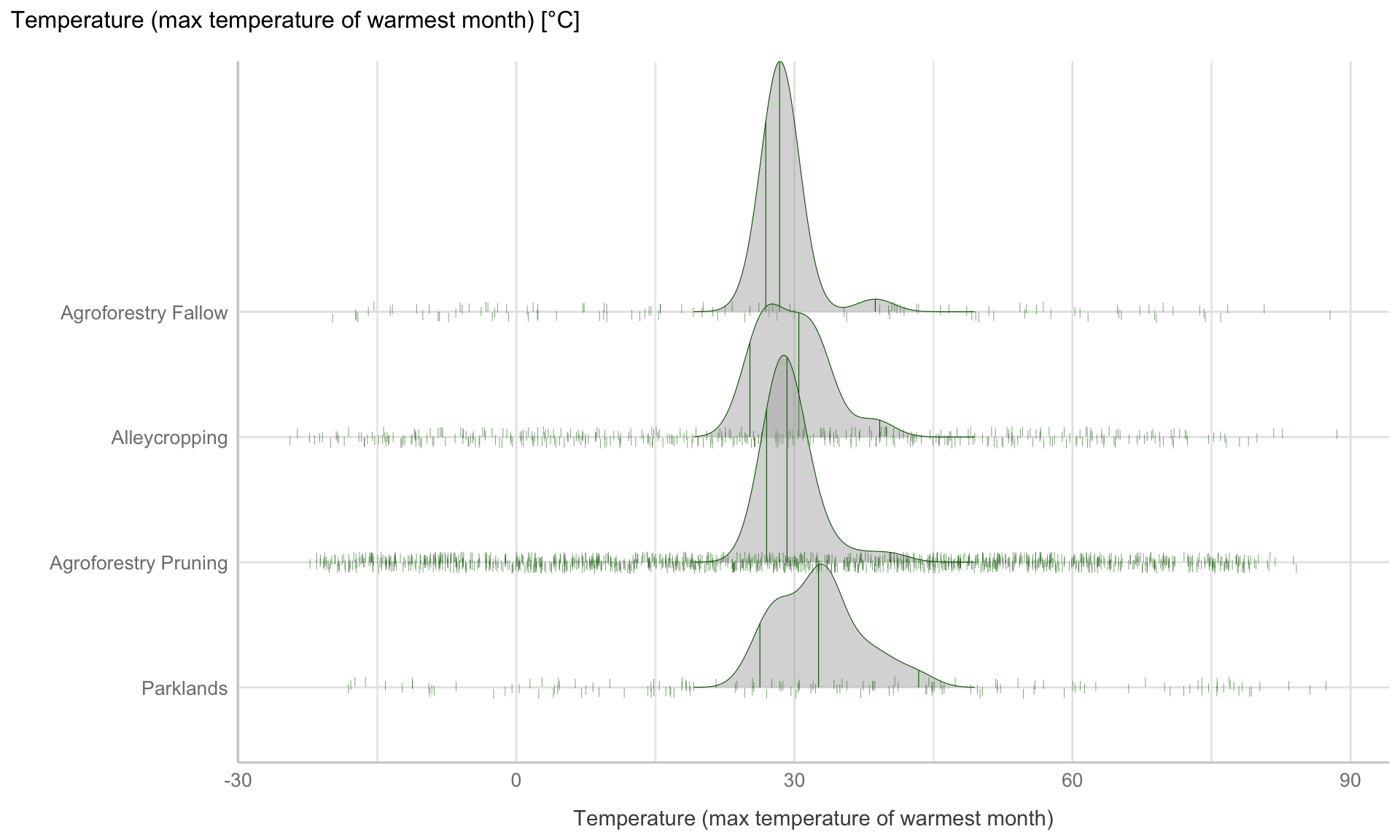
Figure 11: Ridge line plot : Distribution of temperature of warmest month for the four different agroforestry practices
Using the PrepareERA function
As mentioned above, there are some important pre-processing steps that we need to apply to the data before using the ERAAnalyze function. Luckily the ERAg::PrepareERA function can help us perform these pre-processing steps to the data. These steps include dealing with negative response ratio outcomes and dealing with inverse outcomes, like reversing MeanC (control) and MeanT (treatment) if it happens that MeanC is better than MeanT. The steps performed in ERAg::PrepareERA is:
Negative outcomes: where outcomes have a notable number (e.g. 0.5%) of negative values for the control (MeanC column) or experimental treatment (MeanT column), this is because negative numbers are incompatible with ratios.
Inverse outcomes: when a lower value indicates a better outcome, the outcome values for control (MeanC) and experimental treatment (MeanT) are swapped (excluding economic outcomes).
Alright, Now that we know what the PrepareERA function is doing. Below we are applying the PrepareERA functions to the ERA agroforestry data.
# To avoid common problems of data tables we are going to use the
# data.table::copy() function to add our agroforestry.prac.thresh data
agroforestry.prepared <-
ERAg::PrepareERA(Data = data.table::copy(agroforestry.prac.thresh), Perc.Neg = 0.5, RmNeg = T)
# Have we lost any data?
# dim(agroforestry.prac.thresh) # <- before PrepareERA
# [1] 6361 217
#dim(agroforestry.prepared) # <- after PrepareERA
# [1] 6277 66
| Total number of observations in the dataset | Total number of column features in the dataset | |
|---|---|---|
| Before PrepareERA | 6361 | 217 |
| After PrepareERA | 6277 | 66 |
We have lost some column features from the dataset as expected when PrepareERA is applied. We have also lost a few observations, indicating that there were negative outcomes, that would have caused us an issue in the ERAAnalyze function.
Calculating RR with ERAAnalyze
Now we can perform the analysis to calculate response ratios (RR) of the outcome sub-indicator and practices using the ERAAnalyze function. This will tell us what combination of agroforestry practice and outcome yields the best response ratio. Interesting right? We can ultimately use this information to for example answer a question like: What agroforestry practices are significantly better at increasing Soil Carbon compared to their non-agroforestry counterparts (monoculture)?
Below we are applying the ERAAnalyze function to the agroforestry data
agroforestry.analyzed <-
# pre-processed and prepared ERA agroforestry data
ERAg::ERAAnalyze(agroforestry.prepared,
# extreme outliers are detected and removed for grouping variables
rmOut = TRUE,
# statistics will be compiled for each combination of these variables: Sub-Indicator (Outcome) and Practices
Aggregate.By = c("Out.SubInd","PrName"),
# the number of decimal places in the output
ROUND = 5,
# Fast = FALSE: lmer and lm models are used to estimate means, errors and significance
Fast = FALSE)
# Ignore these warning messages: boundary (singular) fit: see ?isSingular
Lets have a look at the various columns that come out of the ERAAnalyze function:
Show code
[1] "Out.SubInd" "PrName" "Observations"
[4] "Studies" "Sites" "RR.Shapiro.Sig"
[7] "RR" "RR.median" "RR.var"
[10] "RR.se" "RR.Quantiles0.25" "PC.Shapiro.Sig"
[13] "PC" "PC.median" "PC.se"
[16] "PC.var" "PC.Quantiles0.25" "Units"
[19] "Model" "MeanT.Obs" "MeanT"
[22] "MeanT.se" "MeanC.Obs" "MeanC"
[25] "MeanC.se" "RR.t value" "RR.Pr(>|t|)"
[28] "RR.Sigma2" "PC.t value" "PC.Pr(>|t|)"
[31] "PC.Sigma2" "RR.pc.se.low" "RR.pc"
[34] "RR.pc.se.high" "RR.pc.jen.low" "RR.pc.jen"
[37] "RR.pc.jen.high" "PC.pc.se.low" "PC.pc"
[40] "PC.pc.se.high" Note: For a detailed description of the ERAAnalyze output type ?ERAg::ERAAnalyze in R - if you have ERAg package installed.
Because we have relatively sparse number of observation for some combinations of our grouping variables we can use the data availability fields to filter the results based on a minimum number of studies. It is always good to have more that one study to contribute to the RR. The number of studies one wish to set as threshold depends on the specific analysis and the power and robustness one wish to obtain. Here we are going to set the combinations that meet a minimum data requirement based on number of studies.
Threshold: Minimum number of studies
Luckily we are working with a highly aggregated dataset with fairly large amounts of observations from plenty of studies. So we can specify a threshold of a minimum of 2 studies.
agroforestry.analyzed.thresh <- agroforestry.analyzed[Studies >= 2]
# Have we lost any data?
dim(agroforestry.analyzed) # <- before Studies >= 2
# [1] 166 40
dim(agroforestry.analyzed.thresh) # <- after Studies >= 2
# [1] 74 40
| Total number of Sub-Outcome and Practice combinations | Total number of column features | |
|---|---|---|
| Before Studies >= 2 | 166 | 40 |
| After Studies >= 2 | 74 | 40 |
Note: Again, This is a rather arbitrary number that depends on what exactly one wish to analyse and how much data is available after the ERAAnalyze.
Yes we did lose some of our data but in this way we are having a relatively more robust analysis outcomes. Hence we can make more generalisable conclusions, and this is especially important for this case - as we wish to look at the general impact of agroforestry accros crop and biomass yields for annual crops.
The potential and actual threshold of number of studies is very limited by the total amount of data available. In the case of very dis-aggregated data (e.g. Effect of the Practice “Agroforestry Pruning under Reduced Tillage with Water Harvesting” on the Outcome “Labour Hours per Person”) there are typically very little data available, possibly only from a few number of studies, hence we have no choice but to accept RR derived from relatively poor quality data.
RR of agroforestry practices
Note: A response ratio (RR) is simply the natural log of the ratio of the experimental outcome to the control outcome. If maize yields with planting basins are 1.5 Mg/ha and without them are 1.1 Mg/ha the response ratio is log(1.5/1.1) = 0.310. RRs greater than zero indicate the experimental treatment is better than the control and vice-versa for RRs less than zero.
Let’s now have a look at the response ratios of the agroforestry data. While remembering the most important information generated from the ERAAnalyze function. The highlighted are the most important to remember as these are what we are going to use to evaluate the response ratio performance of agroforestry practices.
The test results on RR for practices are presented in the “t value,” “Pr(>|t|)” and “Sigma2” columns, where “Pr(>|t|)” indicates significance.
A Shapiro-Wilks test of normality is applied to RRs. If a value in the RR.Shapiro.Sig field is <0.05 then the data for that row are statistically non-normal therefore estimates of central tendency and tests results may be unreliable.
Model = type of model that was applied to data, NA = no model was applied.
RR.t value = t statistic from RR model
RR.Pr(>|t|) = probability that the outcome is not equal to zero from RR model
RR.Sigma2 = RR model sigma2 - quantifies how much the responses (y) vary around the (unknown) mean
PC.Sigma2 = PC model sigma2
RR.pc = % change based on RR (100 x exp(RR) - 100)
RR.pc.se.low = lower standard error confidence interval of % change based on RR
RR.pc.se.high = upper standard error confidence interval of % change based on RR
RR.pc.jen = % change based on RR with correction for Jensen’s inequality (100 x exp(RR+RR.var/2) - 100)
RR.pc.jen.low = lower standard error confidence interval of % change based on RR with correction for Jensen’s inequality
RR.pc.jen.high = upper standard error confidence interval of % change based on RR with correction for Jensen’s inequality
Note: For easier interpretation mean RR and RR +/- standard errors are back-transformed and converted into percentage change (e.g., a ratio of 1.1 = a 10% increase). The RR.pc. columns back-transform log ratios using the exponent. The RR.pc.jen. columns back-transform log ratios using the exponent with a correction for the Jensen’s inequality Tanadini and Mehrabi (2017). We are going to use these back-transformed and corrected response ratios in percent units from RR.pc.jen, as these are easier to interpret.
agroforestry.analyzed.thresh <- readr::read_csv(here::here("ERAAnalyze_OUTPUT", "agroforestry.analyzed.thresh.csv"))
# Selecting the columns of interest in the ERAAnalyze function output
agroforestry.analyzed.rr <- agroforestry.analyzed.thresh %>%
dplyr::select(Out.SubInd,
PrName,
RR.Shapiro.Sig,
RR,
RR.Sigma2,
PC.Sigma2,
RR.pc.jen,
RR.pc.jen.high,
RR.pc.jen.low,
starts_with("RR.t value"),
starts_with("RR.Pr(>|t|)")) %>%
# Renaming "RR.t value" into "RR.t.value"
dplyr::rename_(RR.t.value = 10) %>%
# Renaming "RR.Pr(>|t|)" into something a little easier "RR.Pr.t"
dplyr::rename_(RR.Pr.t = 11) %>%
dplyr::relocate(Out.SubInd, PrName, RR.t.value, RR.Sigma2, RR.Pr.t, RR.pc.jen, RR.Shapiro.Sig) %>%
# sorting the table in descending order for the columns "RR.t.value" and "RR.pc.jen"
dplyr::arrange(desc(RR.t.value, RR.pc.jen))
# Viewing it in a table
rmarkdown::paged_table(agroforestry.analyzed.rr)
Out.SubInd <chr> | |
|---|---|
| Soil Organic Carbon | |
| Crop Yield | |
| Biomass Yield | |
| Crop Yield | |
| Crop Yield | |
| Crop Yield | |
| Soil Organic Carbon | |
| Crop Yield | |
| Crop Yield | |
| Soil Organic Matter |
Back to the agroforestry data: Density distributions
Before we get into the results of the ERAAnalyze function, let us first check the density distribution for the most data rich combination of Outcome and Practice to get a better idea of the data. This is the Outcome crop and biomass yield and the Practice “Agroforestry Pruning.” Hence, we are going to look at the density distribution of the RR of the Crop Yield variable.
Show code
# Reading/loading the agroforestry.prepared dataset
agroforestry.prepared <- readRDS(here::here("ERAAnalyze_OUTPUT", "agroforestry.prepared.RDS"))
agroforestry.prepared.yi <-
agroforestry.prepared %>%
dplyr::filter(Out.SubInd == "Crop Yield" |
Out.SubInd == "Biomass Yield") %>%
rationalize(yi) %>%
drop_na(yi) %>%
ggpubr::gghistogram(
x = "yi",
y = "..count..",
add = "mean",
rug = TRUE,
color = "#E7B800",
add_density = TRUE,
bins = 40) +
ggtitle("Histogram and density distribution for crop and biomass yields") +
xlab("Relative crop and biomass yield") +
ylab("Counts of identical cases") +
theme_lucid(plot.title.size = 25,
axis.text.size = 15,
legend.title.size = 20,
legend.text.size = 20,
axis.title.size = 20)
agroforestry.prepared.yi
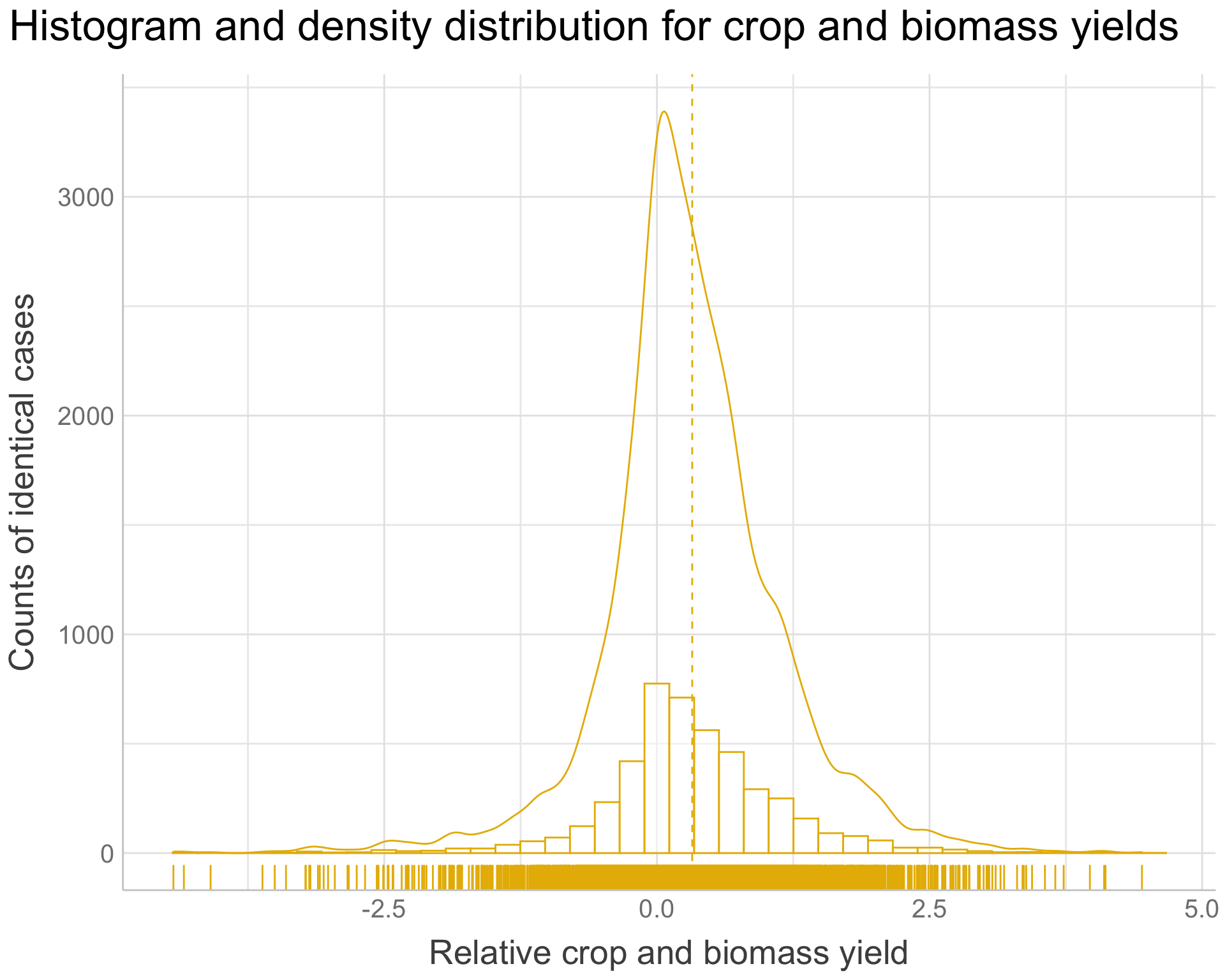
Figure 12: Histogram and density distribution plot for RR of crop and biomass yield
We are observing that:
The RR of crop and biomass yield has a fairly good resemblance to an ideal normal distribution centred around the mean.
The mean of the RR of crop and biomass yield is skewed to the right indicating an overall positive effect of agroforestry on crop and biomass yield , even though we do see a substantial amount of cases with negative RR for crop and biomass yield . Thus, in certain situations there will be an overall negative effect of agroforestry compared to non-agroforestry and we cannot conclude that agroforestry pruning practices is better than non-agroforestry in all situations.
We can make a similar plot for the proportions of MeanC/MeanT with the agroforestry data of Outcome crop and biomass yield and the Practice “Agroforestry Pruning”:
Show code
agroforestry.prepared.meanc.menat <-
agroforestry.prepared %>%
dplyr::filter(Out.SubInd == "Crop Yield" |
Out.SubInd == "Biomass Yield") %>%
dplyr::mutate(Proportion = MeanC/MeanT) %>%
ggpubr::gghistogram(
x = "Proportion",
y = "..count..",
add = "mean",
rug = TRUE,
color = "#66C2A5",
add_density = TRUE,
bins = 200) +
ggtitle("Proportions of MeanC/MeanT for crop and biomass yield") +
xlab("Proportion [MeanC/MeanT]") +
ylab("Counts of identical cases") +
theme_lucid(plot.title.size = 25,
axis.text.size = 15,
legend.title.size = 20,
legend.text.size = 20,
axis.title.size = 20)
agroforestry.prepared.meanc.menat
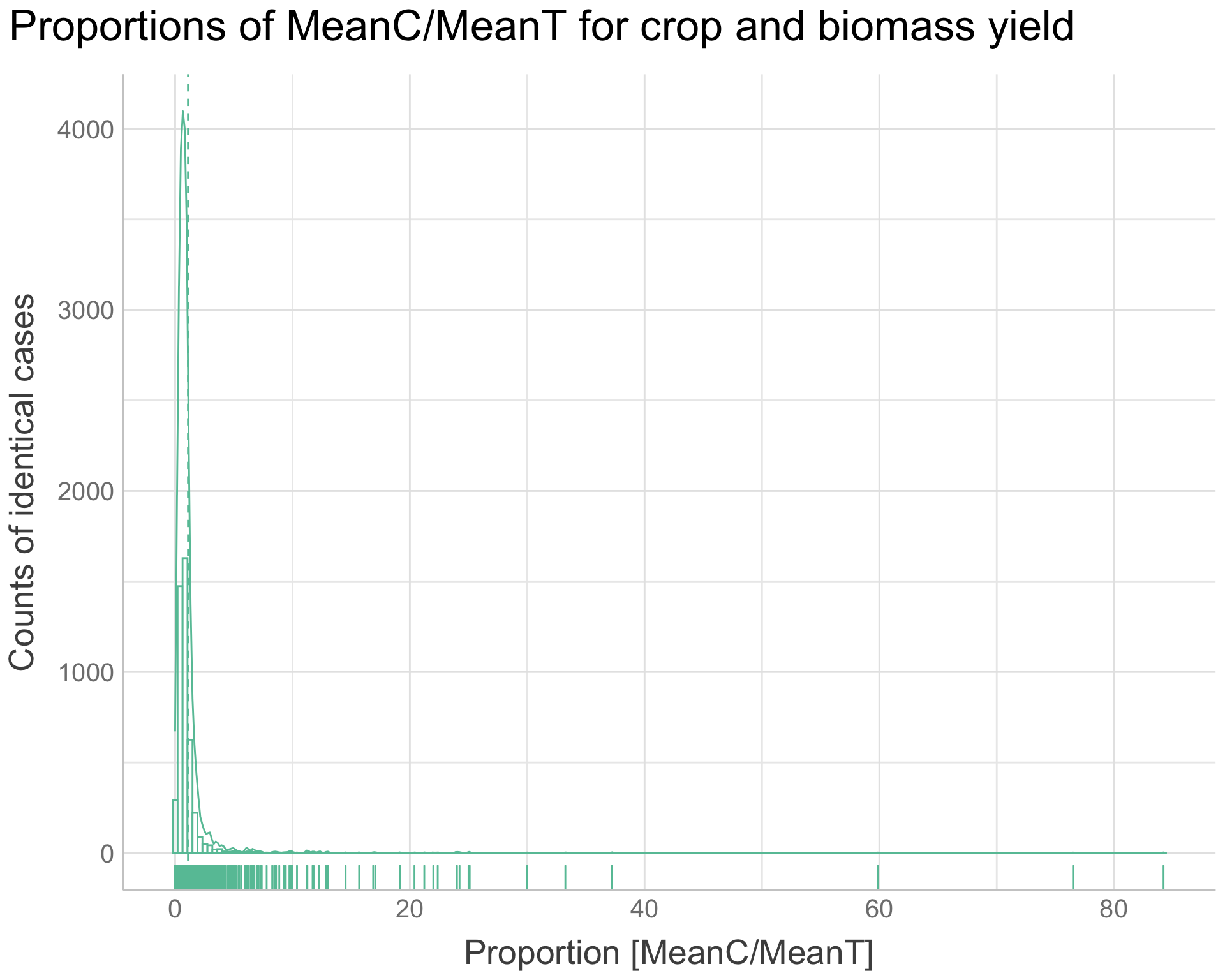
Figure 13: Proportions of MeanC/MeanT for crop and biomass yield
This gives us a clear indication of why the log-transformation is important to our RR outcome. If not we would get this long tail on the distribution.
Results of ERAAnalyze
Great! Now we can interpret the results coming from ERAAnalyze and ask ourself some pretty interesting questions. For example, does practising agroforestry result in higher crop yields compared to non-agroforestry? Or can we identify any significant positive contribution on soil organic carbon when agroforestry is practised?
What is the proportional effect of different agroforestry practices on response ratios (RR)? Or said in another way. Which agroforestry practices are performing best and which are performing worse?
To visually summarise results that can answer this question a simple bar plot is great! If we want to make a simple bar plot that shows the proportional impact on RR for the different agroforestry practices the best thing we can do is to use the back-transformed RR in percentage, that have been corrected for Jensen’s inequality - found in the column “RR.pc.jen.”
However, before we proceed to actually making the bar plot of proportional effects on RR, we have to deal with two important issues:
Outliers
Missing values
Omitting outliers from the analysed data
Omitting RR and RR.pc.jen outliers from the analysed agroforestry data
Identifiyng outliers
Identifiyng outliers in RR and RR.pc.jen values
We will use a combined outlier detection method as suggested by Evgeni Chasnovski on his website, Question Flow. We are going to perform the combined outlier detection method by applying functions from the packages “dplyr” and “ruler,” to the analysed agroforestry data. This method is based on combining different outlier detection techniques to identify rows which are “strong outliers” and which might by considered outliers based on several methods. The combined outlier detection method can be divided in steps of:
- Defining outlier detection method
- Definition of group-based non-outlier rows
- Exposure: Non-outlier rows
- Combination: Using the combined outlier detection score for certain row
- Identify rows with strong outliers
- Illustrating strong outliers in data with visualisation
Defining outlier detection methods
Show code
# Z-score
isnt_out_z <- function(x, thres = 3, na.rm = TRUE) {
abs(x - mean(x, na.rm = na.rm)) <= thres * sd(x, na.rm = na.rm)
}
# Z-score with MAD
isnt_out_mad <- function(x, thres = 3, na.rm = TRUE) {
abs(x - median(x, na.rm = na.rm)) <= thres * mad(x, na.rm = na.rm)
}
# Tukeys's fences
isnt_out_tukey <- function(x, k = 1.5, na.rm = TRUE) {
quar <- quantile(x, probs = c(0.25, 0.75), na.rm = na.rm)
iqr <- diff(quar)
(quar[1] - k * iqr <= x) & (x <= quar[2] + k * iqr)
}
# Mahalanobis distance
maha_dist <- . %>% select_if(is.numeric) %>%
mahalanobis(center = colMeans(.), cov = cov(.))
isnt_out_maha <- function(tbl, isnt_out_f, ...) {
tbl %>% maha_dist() %>% isnt_out_f(...)
}
# Combined methodology
isnt_out_funs <- funs(
z = isnt_out_z,
mad = isnt_out_mad,
tukey = isnt_out_tukey
)
# Warning: `funs()` was deprecated in dplyr 0.8.0.
# Please use a list of either functions or lambdas:
#
# # Simple named list:
# list(mean = mean, median = median)
#
# # Auto named with `tibble::lst()`:
# tibble::lst(mean, median)
#
# # Using lambdas
# list(~ mean(., trim = .2), ~ median(., na.rm = TRUE))
# This warning is displayed once every 8 hours.
# Call `lifecycle::last_warnings()` to see where this warning was generated.
The resulting function (isnt_out_funs) of the combined methods has outputs for three methods (Z-score, Z-score with MAD and Tukey’s fences). Their names are considered as method names.
Note: that all listed approached depend on the choice of the univariate outlier detection method. We will use all three previously listed univariate techniques.
Implementation: Column based non-outlier rows
For the “agroforestry.analyzed” dataset rules for column based non-outlier rows can be defined based on 7 numeric columns and 3 presented univariate detection methods. There is a convenient way of computing all them at once using scoped variant of dplyr::transmute():
Show code
agroforestry.analyzed.non.out <- agroforestry.analyzed %>%
transmute_if(is.numeric, isnt_out_funs)
rmarkdown::paged_table(agroforestry.analyzed.non.out)
Observations_z <lgl> | Studies_z <lgl> | Sites_z <lgl> | RR.Shapiro.Sig_z <lgl> | RR_z <lgl> | RR.median_z <lgl> | |
|---|---|---|---|---|---|---|
| TRUE | TRUE | TRUE | TRUE | TRUE | TRUE | |
| TRUE | TRUE | TRUE | TRUE | TRUE | TRUE | |
| TRUE | TRUE | TRUE | TRUE | TRUE | TRUE | |
| TRUE | TRUE | TRUE | TRUE | TRUE | TRUE | |
| TRUE | TRUE | TRUE | TRUE | TRUE | TRUE | |
| TRUE | TRUE | TRUE | TRUE | TRUE | TRUE | |
| TRUE | FALSE | FALSE | TRUE | TRUE | TRUE | |
| TRUE | TRUE | TRUE | TRUE | TRUE | TRUE | |
| FALSE | FALSE | FALSE | TRUE | TRUE | TRUE | |
| FALSE | FALSE | FALSE | TRUE | TRUE | TRUE |
The output of the code above gives us something like a logical Matrix with “TRUE”/“FALSE” statements for each of the column features for unique groups of Practices (PrName) and Outcomes (Out.SubInd). This result has outputs for all the methods applied to the 166 groups. Their names are of the form
Implementation: Mahalanobis based non-outlier rows
To define non-outlier rows based on Mahalanobis distance one can apply univariate method for distances computed for some subset of numeric columns. To simplify a little bit, we will choose one “subset” with all numeric columns and all listed methods.
Implementation: Group based non-outlier rows
Definition of non-outlier rows based on groupings depends on a group summary function and univariate outlier detection methods. As grouping column we will choose the two non-numeric columns Practices (PrName) and Outcomes (Out.SubInd), and unite them into one called “PrName_Out.SubInd” - this will make it easier for our later imputation of non-outlier rows.
agrofor.data_tbl <- agroforestry.analyzed %>%
unite(col = "PrName_Out.SubInd", PrName, Out.SubInd)
compute_group_non_outliers <- . %>%
# Compute per group mean values of columns
group_by(PrName_Out.SubInd) %>%
summarise_if(is.numeric, mean) %>%
ungroup() %>%
# Detect outliers among groups
mutate_if(is.numeric, isnt_out_funs) %>%
# Remove unnecessary columns
select_if(Negate(is.numeric))
rmarkdown::paged_table(agrofor.data_tbl %>% compute_group_non_outliers())
PrName_Out.SubInd <chr> | |
|---|---|
| Agroforestry Fallow_Biomass Yield | |
| Agroforestry Fallow_Crop Yield | |
| Agroforestry Fallow_Infiltration Rate | |
| Agroforestry Fallow_Soil Nitrogen | |
| Agroforestry Fallow_Water Use Efficiency | |
| Agroforestry Fallow-Agroforestry Pruning_Biomass Yield | |
| Agroforestry Fallow-Agroforestry Pruning_Crop Yield | |
| Agroforestry Fallow-Agroforestry Pruning_Infiltration Rate | |
| Agroforestry Fallow-Agroforestry Pruning_Soil Nitrogen | |
| Agroforestry Fallow-Agroforestry Pruning_Soil Organic Carbon |
The output of the code above gives us something like a logical Matrix with “TRUE”/“FALSE” statements for each of the column features for unique groups of Practices (PrName) and Outcomes (Out.SubInd). This result has outputs for all the methods applied to the 166 groups. Their names are of the form
Exposure Column and Mahalanobis based definition of non-outlier rows can be expressed with row packs and group based - as group packs. This is syntax from the ruler package
row_packs_isnt_out <- ruler::row_packs(
# Non-outliers based on some column
column = . %>% transmute_if(is.numeric, isnt_out_funs),
# Non-outliers based on Mahalanobis distance
maha = . %>% transmute(maha = maha_dist(.)) %>%
transmute_at(vars(maha = maha), isnt_out_funs)
)
group_packs_isnt_out <- ruler::group_packs(
# Non-outliers based on grouping
PrName_Out.SubInd = compute_group_non_outliers,
.group_vars = "PrName_Out.SubInd"
)
Application of all those packs is called exposing process. The result is an exposure from which we can extract tidy data validation report using get_report.
# Don't remove obeyers to compute total number of applied rules
full_report <- agrofor.data_tbl %>%
expose(row_packs_isnt_out, group_packs_isnt_out,
.remove_obeyers = FALSE) %>%
get_report()
used_rules <- full_report %>%
distinct(pack, rule)
breaker_report <- full_report %>%
filter(!(value %in% TRUE))
used.rules contains data about all definitions of non-outlier rows applied to our analysed agroforestry data. They are encoded with combinations of columns pack and rule.
breaker_report contains data about data units that break certain rules. Packs column and maha has actual row numbers of data_tbl listed in id column of report (for rows which should be considered as outliers).
Pack group defines group pack and is represented in breaker_report with id 0. To obtain row outliers based on grouping we need to expand those rows with information about rows in the data that belong to those groups. This can be done using dplyr::left_join() function:
# Defining new dataset with group based breakers
group_breakers <- breaker_report %>%
# Filter PrName_Out.SubInd packs
dplyr::filter(pack == "PrName_Out.SubInd") %>%
# Expand rows by matching PrName_Out.SubInd with its rows
dplyr::select(-id) %>%
dplyr::left_join(y = agrofor.data_tbl %>% transmute(var = PrName_Out.SubInd, id = 1:n()),
by = "var") %>%
dplyr::select(pack, rule, var, id, value)
# Splitting based on our grouping variable PrName_Out.SubInd
outliers <- bind_rows(
breaker_report %>% filter(pack != "PrName_Out.SubInd"),
group_breakers
) %>%
select(pack, rule, id)
# Not all group based definitions resulted with outliers
outliers2 <- outliers %>%
count(pack, rule) %>%
dplyr::filter(pack == "PrName_Out.SubInd") %>%
print(n = Inf)
Input is not proper 'ruler_report' object.# Warning
# Input is not proper 'ruler_report' object.
rmarkdown::paged_table(outliers)
pack <chr> | rule <chr> | id <int> | ||
|---|---|---|---|---|
| column | Observations_z | 9 | ||
| column | Observations_z | 10 | ||
| column | Observations_z | 16 | ||
| column | Studies_z | 7 | ||
| column | Studies_z | 9 | ||
| column | Studies_z | 10 | ||
| column | Studies_z | 16 | ||
| column | Sites_z | 7 | ||
| column | Sites_z | 9 | ||
| column | Sites_z | 10 |
The output of the code above gives us a tibble called “outliers” which contains data about outlier rows.
- Combination of columns pack and rule defines non-outlier/outlier definition approach.
- Column id defines the row number of the input dataframe (our agroforestry data) that should be considered an outlier based on the definition.
outlier_score <- outliers %>%
group_by(id) %>%
# nrow(used_rules) equals total number of applied methods
summarise(score = n() / nrow(used_rules))
# Top 10 outliers
outlier_score %>% arrange(desc(score)) %>% dplyr::slice(1:10)
# A tibble: 10 × 2
id score
<int> <dbl>
1 33 0.700
2 60 0.633
3 59 0.585
4 35 0.575
5 22 0.565
6 98 0.546
7 48 0.527
8 85 0.527
9 97 0.527
10 41 0.507Given the dataframe “outliers,” one can do whatever he/she wants to identify outliers. Here we will use the basic combination approach based on average scores. You can see in the table above, where a random sample of 10 rows is shown, that all have an “outlier score.” We can use this score to further process the data so that we remove outliers that are above a determined combined outlier score. Combined outlier detection score for certain row can be defined as share of applied methods that tagged it as outlier. Alternatively one can define it just as number of those methods as it will only change absolute value of the result and not the order.
Next we will use the combined outlier detection score to remove observations from our analysed agroforestry dataframe (agroforestry.analyzed), by setting a threshold to 0.3 and all observations above 0.3 will be removed.
Note: that the threshold here is again arbitrary and depends on the nature of the data, and the total availability of data. In our case we have 166 rows, from each unique combination of Outcome (Out.SubInd) and Practice (PrName). Hence we do not want to set the threshold too low as that would remove too many observations.
agroforestry.outlier.id <- agroforestry.analyzed %>%
mutate(id = 1:n()) %>%
left_join(y = outlier_score, by = "id") %>%
mutate(
score = coalesce(score, 0),
is_out = if_else(score > 0.40, "Outlier", "Not outlier")
)
# Total number of outliers
sum(agroforestry.outlier.id$score > 0.40)
[1] 36 # Threshold: score > 0.20 giving 115 removed
# Threshold: score > 0.35 giving 53 removed
# Threshold: score > 0.40 giving 36 removed
# Threshold: score > 0.50 giving 13 removed
Visualising and assessing outliers
We can get a large variety of information out of this outlier analysis. Here we are going to look at the outliers for the “original” RR and percentage corrected RR.pc.jen as illustrated by: 1) Practices, 2) Outcomes and 3) Number of studies.
Show code
# Creating the functions that will generate the plots
# Plot 1: Strong RR and RR.jpc.jen outliers by practices
agroforestry.analyzed.outliers.plot.PrName <- function(tbl, x, y) {
tbl %>%
arrange(is_out) %>%
ggplot(aes_string(x, y, colour = "is_out")) +
# applying a little jittering so points are distinguishable
geom_jitter(size=3, alpha=0.8, width = 0.8, height = 0.1) +
#facet_wrap(facets = facet_var) +
scale_colour_manual(values = c("#66C2A5", "black")) +
guides(colour = guide_legend(title = NULL,
override.aes = list(size = 5))) +
#labs(title = paste0("Strong outliers illustration by practices")) +
theme_lucid(plot.title.size = 25,
axis.text.size = 16,
legend.title.size = 20,
legend.text.size = 20,
axis.title.size = 20) +
theme(legend.position = "top",
legend.text = element_text(size = 10),
axis.text.x = element_text(angle = 0, hjust=1))
}
# Plot 2: Strong RR and RR.jpc.jen outliers by outcomes
agroforestry.analyzed.outliers.plot.Outcome <- function(tbl, x, y) {
tbl %>%
arrange(is_out) %>%
ggplot(aes_string(x, y, colour = "is_out")) +
# applying a little jittering so points are distinguishable
geom_jitter(size=3, alpha=0.8, width = 0.8, height = 0.1) +
#facet_wrap(facets = facet_var) +
scale_colour_manual(values = c("#66C2A5", "black")) +
guides(colour = guide_legend(title = NULL,
override.aes = list(size = 5))) +
# labs(title = paste0("Strong outliers illustration by outcome")) +
theme_lucid(plot.title.size = 25,
axis.text.size = 16,
legend.title.size = 20,
legend.text.size = 20,
axis.title.size = 20) +
theme(legend.position = "top",
legend.text = element_text(size = 10),
axis.text.x = element_text(angle = 0, hjust=1))
}
# Plot 3: Strong RR and RR.jpc.jen outliers by number of studies
agroforestry.analyzed.outliers.plot.NoStudies <- function(tbl, x, y) {
tbl %>%
arrange(is_out) %>%
ggplot(aes_string(x, y, colour = "is_out")) +
# applying a little jittering so points are distinguishable
geom_jitter(size=3, alpha=0.8, width = 0.8, height = 0.1) +
#facet_wrap(facets = facet_var) +
scale_colour_manual(values = c("#66C2A5", "black")) +
guides(colour = guide_legend(title = NULL,
override.aes = list(size = 5))) +
# labs(title = paste0("Strong outliers illustration by number of studies")) +
theme_lucid(plot.title.size = 25,
axis.text.size = 16,
legend.title.size = 20,
legend.text.size = 20,
axis.title.size = 20) +
theme(legend.position = "top",
legend.text = element_text(size = 10),
axis.text.x = element_text(angle = 0, hjust=1)) +
coord_trans(y="log2")
}
Strong RR and RR.jpc.jen outliers by practices
Show code
plot_PrName_RR.pc.jen <- agroforestry.outlier.id %>% agroforestry.analyzed.outliers.plot.PrName("RR.pc.jen", "PrName")
plot_PrName_RR <- agroforestry.outlier.id %>% agroforestry.analyzed.outliers.plot.PrName("RR", "PrName")
plot_PrName <- ggarrange(plot_PrName_RR.pc.jen, plot_PrName_RR + rremove("y.text") + rremove("ylab"),
labels = c("A)", "B)"),
ncol = 2, nrow = 1)
annotate_figure(plot_PrName,
top = text_grob("Strong outliers illustration by practices \n ",
color = "black", face = "bold", size = 30))
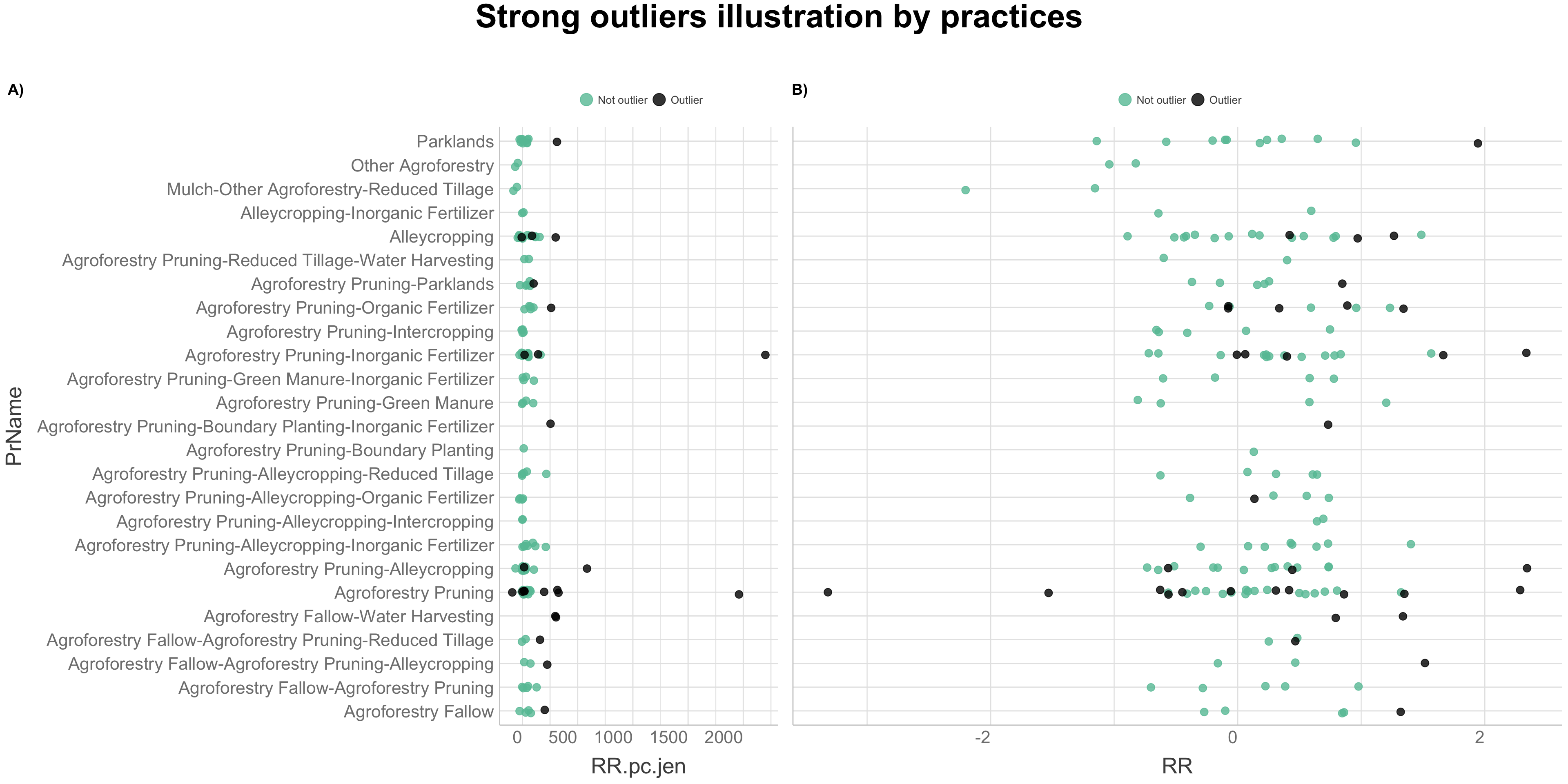
Figure 14: Strong outliers illustration by practices
Strong RR and RR.jpc.jen outliers by outcomes
Show code
plot_Outcome_RR.pc.jen <- agroforestry.outlier.id %>% agroforestry.analyzed.outliers.plot.Outcome("RR.pc.jen", "Out.SubInd")
plot_Outcome_RR <- agroforestry.outlier.id %>% agroforestry.analyzed.outliers.plot.Outcome("RR", "Out.SubInd")
plot_Outcome <- ggarrange(plot_Outcome_RR.pc.jen, plot_Outcome_RR + rremove("y.text") + rremove("ylab"),
labels = c("A)", "B)"),
ncol = 2, nrow = 1)
annotate_figure(plot_Outcome,
top = text_grob("Strong outliers illustration by outcomes \n ",
color = "black", face = "bold", size = 30))
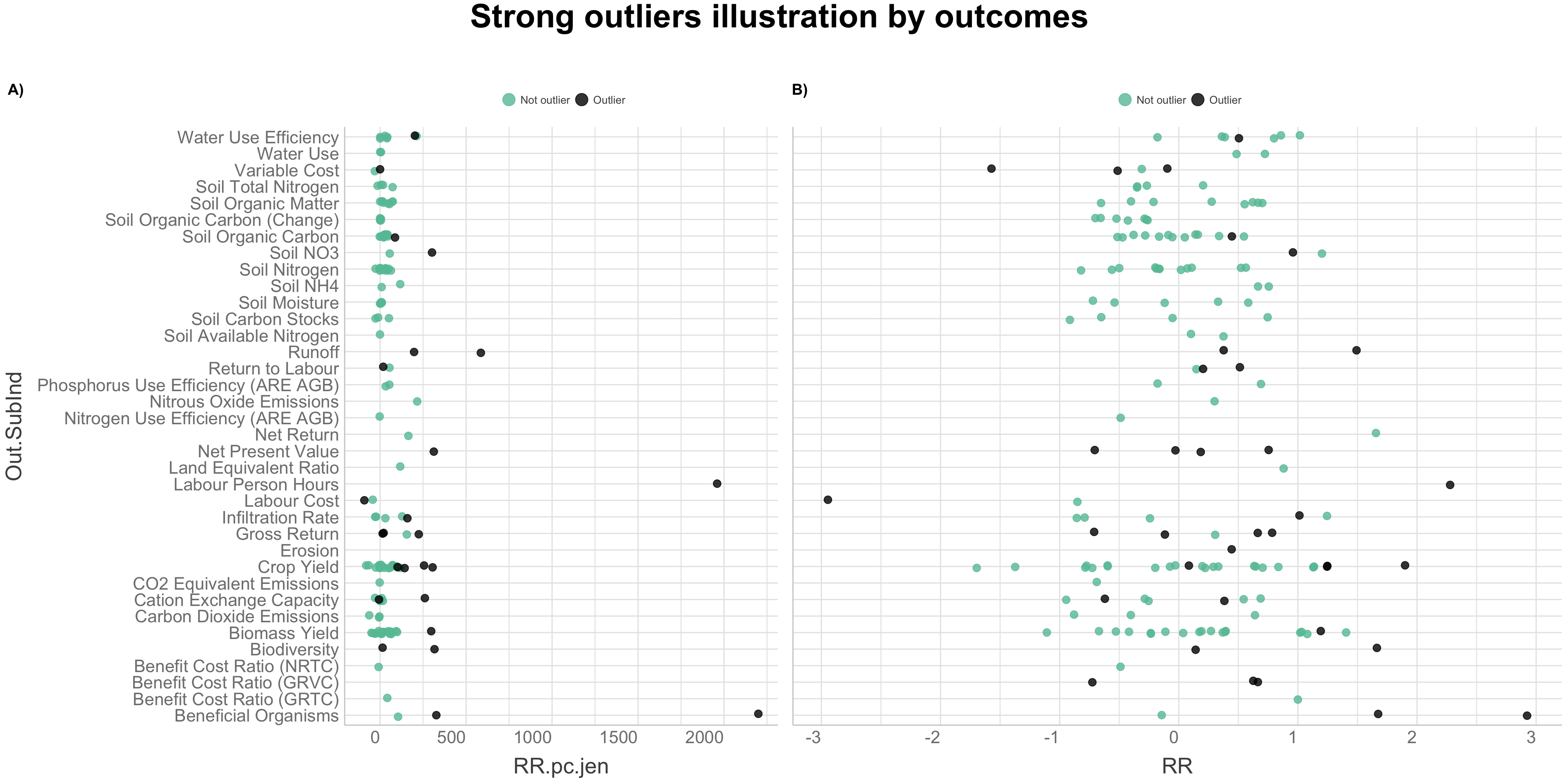
Figure 15: Strong outliers illustration by practices
Strong RR and RR.jpc.jen outliers by number of studies
Show code
plot_Studies_RR.pc.jen <- agroforestry.outlier.id %>% agroforestry.analyzed.outliers.plot.NoStudies("RR.pc.jen", "Studies")
plot_Studies_RR <- agroforestry.outlier.id %>% agroforestry.analyzed.outliers.plot.NoStudies("RR", "Studies")
plot_Studies <- ggarrange(plot_Studies_RR.pc.jen, plot_Studies_RR + rremove("y.text") + rremove("ylab"),
labels = c("A)", "B)"),
ncol = 2, nrow = 1)
annotate_figure(plot_Studies,
top = text_grob("Strong outliers illustration by number of studies \n ",
color = "black", face = "bold", size = 30))
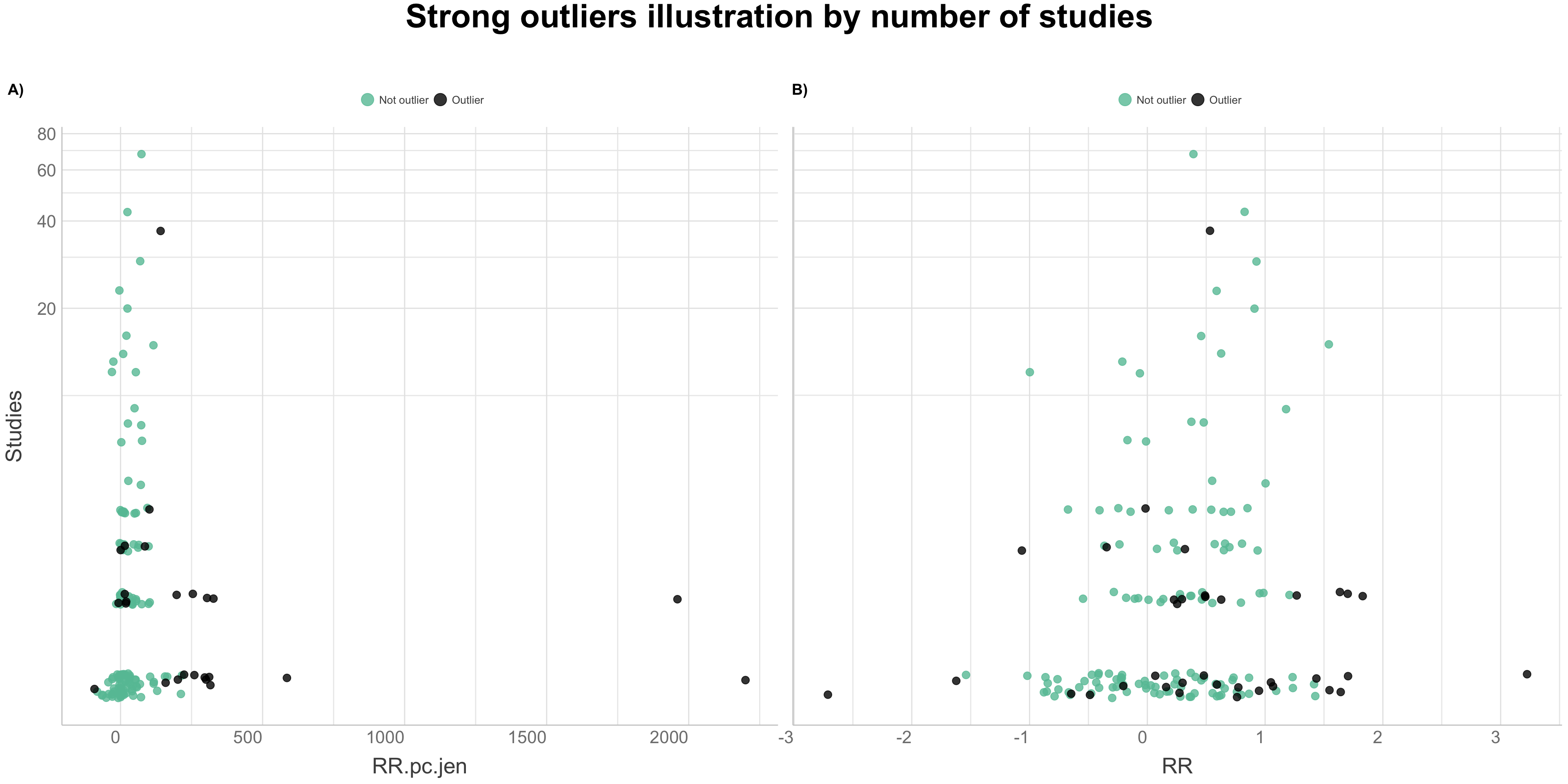
Figure 16: Strong outliers illustration by practices
Groups of Practices and Outcomes with most outliers
We can also look at the individual agroforestry practices and or outcomes (and combinations) to see witch ones have most outliers. Let us now look at what groups of our analysed agroforestry data is most prone to have outliers. We will use the dataset with breaker reports, that we prepared earlier.
Show code
Input is not proper 'ruler_report' object.Show code
rmarkdown::paged_table(agroforestry_breaker.report)
var <chr> | |
|---|---|
| Agroforestry Pruning_Labour Person Hours | |
| Agroforestry Pruning-Inorganic Fertilizer_Beneficial Organisms | |
| Agroforestry Pruning_Biodiversity | |
| Agroforestry Pruning_Variable Cost | |
| Agroforestry Pruning_Beneficial Organisms | |
| Agroforestry Pruning_Erosion | |
| Agroforestry Pruning_Return to Labour | |
| Agroforestry Pruning-Alleycropping_Runoff | |
| Agroforestry Pruning-Organic Fertilizer_Cation Exchange Capacity | |
| Agroforestry Pruning-Inorganic Fertilizer_Variable Cost |
As it could be expected, “Agroforestry Pruning” is among majority of top breaker groups, meaning that this group of agroforestry practices tend to have most outliers. This is expected since most studies, and most data is from this agroforestry practice group. Feel free to go through the groups yourself.
Using only basic outlier detection methods one can achieve insightful results by combining them. Observations which are tagged as outlier by more than some threshold number of methods might be named as “strong outliers.” Those should be considered as outliers based on the whole data rather then on separate features.
Next we are going to exclude outcome-practice combinations with extreme outliers.
Omitting outliers
Omitting outliers in RR and RR.pc.jen columns
We are going to exclude the outcome-practice combinations with extreme outliers using the extreme outlier removal method explained earlier, see section Using the ERAAnalyze function. With this approach all observations that are found beyond 3 * IQR, will be removed. Let us first have a look at the analysed agroforestry data that have been sorted based on RR.pc.jen.
Show code
rmarkdown::paged_table(
agroforestry.analyzed %>%
relocate(PrName, Out.SubInd, RR, RR.se, RR.pc.jen, RR.pc.jen.low, RR.pc.jen.high) %>%
arrange(desc(RR.pc.jen), 10)
)
PrName <chr> | Out.SubInd <chr> | RR <dbl> | RR.se <dbl> | RR.pc.jen <dbl> | RR.pc.jen.low <dbl> | |
|---|---|---|---|---|---|---|
| Agroforestry Pruning | Erosion | 0.64256 | 0.55725 | Inf | NA | |
| Agroforestry Pruning-Inorganic Fertilizer | Beneficial Organisms | 3.03156 | 0.37196 | 2199.61084 | 1485.30990 | |
| Agroforestry Pruning | Labour Person Hours | 1.79849 | 0.46449 | 1960.34916 | NA | |
| Agroforestry Pruning-Alleycropping | Runoff | 1.87892 | 0.42875 | 585.42918 | 346.43561 | |
| Agroforestry Pruning | Beneficial Organisms | 1.17269 | 0.43090 | 326.82333 | 177.40268 | |
| Agroforestry Pruning | Biodiversity | 1.16253 | 0.17412 | 316.86641 | NA | |
| Parklands | Net Present Value | 1.36345 | 0.46081 | 312.28101 | 160.05553 | |
| Agroforestry Fallow-Water Harvesting | Crop Yield | 1.35338 | 0.15934 | 304.63288 | 245.03304 | |
| Alleycropping | Soil NO3 | 0.54931 | 0.38842 | 301.61756 | NA | |
| Agroforestry Fallow-Water Harvesting | Biomass Yield | 1.32393 | 0.23377 | 296.92613 | 214.18443 |
Again we see that indeed it is the practice “Agroforestry Pruning” that has the most extreme outliers! However, outliers are also found for many other agroforestry practices. Next, we are going to remove all these cases of outliers using an extreme outliers removal method where values above or below 3∗IQR (interquartile range) are removed.
Show code
# Using the agroforestry.analyzed data, straight out from the ERAAnalyze function
agroforestry.analyzed.clean <- agroforestry.analyzed %>%
# rationalize from the hablar package transforms all numeric elements to be rational values or NA, thus removes all NaN,Inf and replaces them with NA.
rationalize(RR, RR.se, RR.pc.jen, RR.pc.jen.low, RR.pc.jen.high) %>%
# drop_na from dplyr package is dropping rows containing missing values
drop_na(RR, RR.se, RR.pc.jen, RR.pc.jen.low, RR.pc.jen.high)
# Defining quantile as 0.25 and 0.75 range
Q.RR.pc.jen <- quantile(agroforestry.analyzed.clean$RR.pc.jen, probs=c(0.25, 0.75), na.rm = TRUE)
# inter quantile range is computed based on RR.pc.jen values in the data
iqr.RR.pc.jen <- IQR(agroforestry.analyzed.clean$RR.pc.jen)
up.RR.pc.jen <- Q.RR.pc.jen[2]+3*iqr.RR.pc.jen # Upper Range
low.RR.pc.jen <- Q.RR.pc.jen[1]-3*iqr.RR.pc.jen # Lower Range
# Subsetting the data observations from agroforestry.analyzed that are within the range - hence excluding outliers
agroforestry.analyzed.rm.outliers <- subset(agroforestry.analyzed.clean, agroforestry.analyzed.clean$RR.pc.jen > (Q.RR.pc.jen[1] - 3*iqr.RR.pc.jen) & agroforestry.analyzed.clean$RR.pc.jen < (Q.RR.pc.jen[2]+3*iqr.RR.pc.jen))
Open the code chunk above if you wish to see the code for removing the extreme outliers in the colums RR and RR.pc.jen.
Let us look at how much data we have lost after outliers have been removed:
dim(agroforestry.analyzed) # <- Before removing NA values
# [1] 166 40
dim(agroforestry.analyzed.clean) # <- Before extreme outlier removal
# [1] 107 40
dim(agroforestry.analyzed.rm.outliers) # <- After extreme outlier removal
# [1] 101 40
table(is.na(agroforestry.analyzed.rm.outliers))
# FALSE TRUE
# 3836 204
sapply(agroforestry.analyzed, function(x) sum(is.na(x)))
We see that we have effectively removed outliers amounting to six observations. We also find that we have around 200 missing values in the observations. Next we are going to perform the median imputation on RR and RR.pc.jen values with missing values. We do this to be able to produce our final bar plot of proportional effect on RR and RR.ps.jen for the different agroforestry practices.
Impute missing values in the analysed data
Mean imputation of missing RR and RR.pc.jen values in the analysed agroforestry data
We will now use a median imputation technique for each of the agroforestry groups, where the median is found across groups of outcome and practice.
Note: the median, like any imputation technique, is statistically a caution and bold move to make and the visual outcome of this comparison should be taken with strong precautions that will limit potential conclusions.
agroforestry.analyzed.imputed <- agroforestry.analyzed.rm.outliers %>%
mutate(impute_RR.pc.jen
= replace(RR.pc.jen,
is.na(RR.pc.jen),
median(RR.pc.jen, na.rm = TRUE))) %>%
mutate(impute_RR.pc.jen.low
= replace(RR.pc.jen.low,
is.na(RR.pc.jen.low),
median(RR.pc.jen.low, na.rm = TRUE))) %>%
mutate(impute_RR.pc.jen.high
= replace(RR.pc.jen.high,
is.na(RR.pc.jen.high),
median(RR.pc.jen.high, na.rm = TRUE)))
dim(agroforestry.analyzed.imputed)
[1] 101 43Show code
agroforestry.analysed.imputed.prname.grouped <- agroforestry.analyzed.imputed %>%
group_by(PrName) %>%
dplyr::summarise(meanRR = mean(RR),
meanRR.se = mean(RR.se),
meanRR.pc.jen = mean(impute_RR.pc.jen),
meanRR.pc.jen.low = mean(impute_RR.pc.jen.low),
meanRR.pc.jen.high = mean(impute_RR.pc.jen.high),
n_obs = Observations,
n_studies = Studies)
dim(agroforestry.analysed.imputed.prname.grouped)
[1] 101 8Show code
# [1] 101 8
# Warning: `summarise()` has grouped output by 'PrName'. You can override using the `.groups` argument.
Show code
agroforestry.analyzed.results.plot <-
agroforestry.analyzed.imputed %>%
dplyr::filter(PrName == "Agroforestry Fallow" |
PrName == "Agroforestry Pruning"|
PrName == "Alleycropping"|
PrName == "Parklands") %>%
arrange(desc(-RR.pc.jen)) %>%
ggpubr::gghistogram(
x = "RR.pc.jen",
y = "..density..",
add = "mean",
add.params = list(linetype = "dashed", size = 1.5),
rug = TRUE,
color = "PrName",
fill = "PrName",
palette = "Set2",
alpha = 0.5,
add_density = TRUE,
bins = 10,
facet.by = "PrName",
scales = "free_y") +
ggtitle("Histogram and density distribution for RR of Crop Yields") +
xlab("RR for Crop Yield") +
ylab("Counts of identical cases of RR values") +
theme_lucid(plot.title.size = 25,
axis.text.size = 15,
legend.title.size = 20,
legend.text.size = 20,
axis.title.size = 20)
agroforestry.analyzed.results.plot
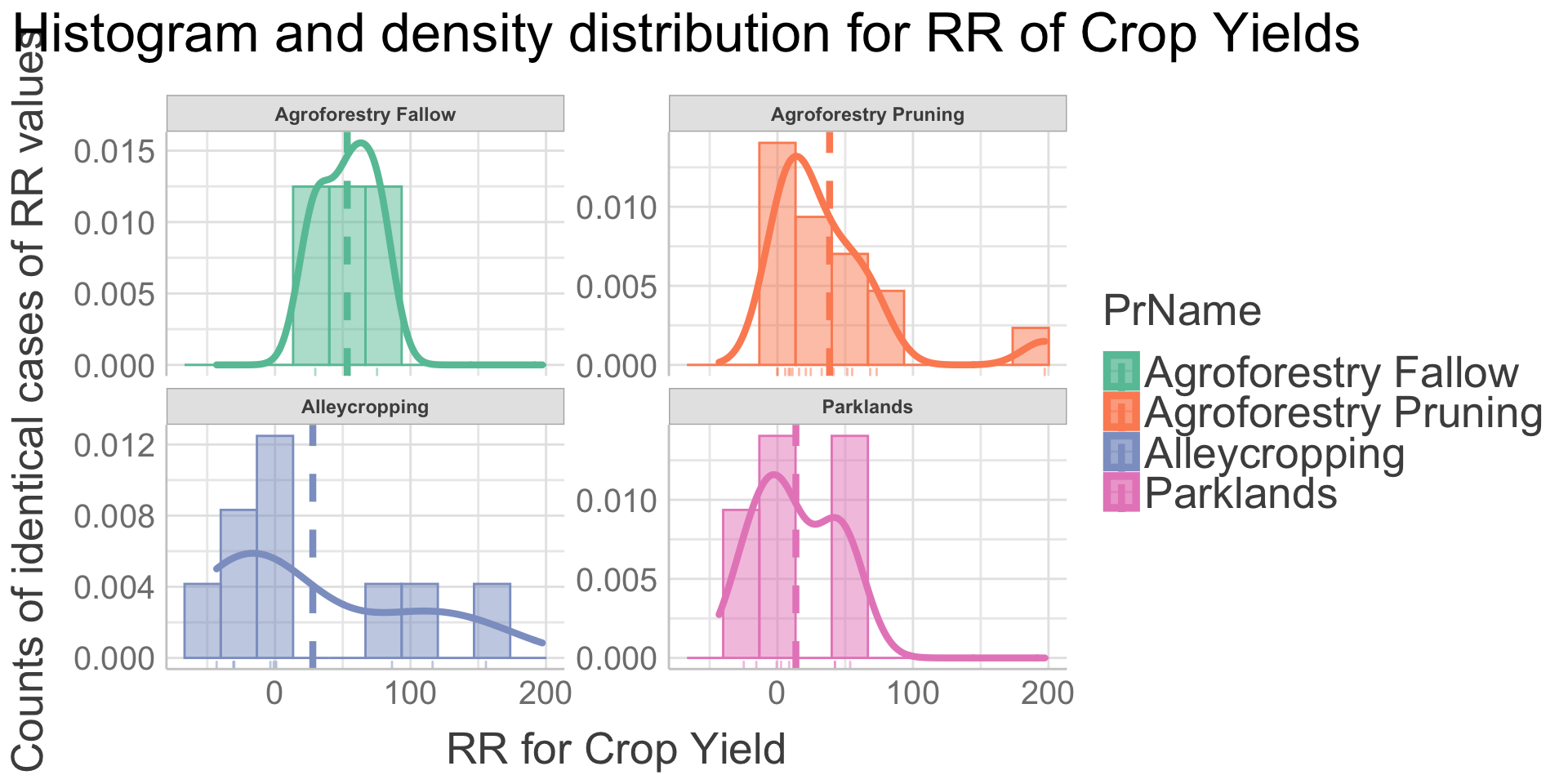
Figure 17: Results of ERAAnalyze for selected agroforestry practices
Show code
agroforestry.analysed.nstudies <- agroforestry.analysed.imputed.prname.grouped %>%
group_by(PrName, meanRR, meanRR.pc.jen) %>% # <- grouping by PrName anf meanRR and meanRR.pc.jen with the assuption that they are all unique
summarise(across(n_obs, list(sum)),
across(n_studies, list(sum))) %>% # <- summarising the number of observations for each combination
mutate(n_obs_label_pos_RR = (n() * 1.5)) %>% # <- making a new column used solely for positioning the n_obs labes on the following bar plots
mutate(n_obs_label_pos_RR.pc.jen = n() * 330) %>%
mutate(n_studies_label_pos_RR = (n() * 1.5)) %>% # <- making a new column used solely for positioning the n_obs labes on the following bar plots
mutate(n_studies_label_pos_RR.pc.jen = n() * 330)
dim(agroforestry.analysed.nstudies)
[1] 24 9Now that we “got rid of the NA values,” we can plot our RR and our RR.pc.jen for each of the different ERA agroforestry practices.
Variations in RR for agroforestry practices
Plotting RR for all agroforestry practices plot in bar plot
Show code
# Defining a custom colours range
nb.cols <- 25
era.af.colors <- colorRampPalette(brewer.pal(8, "Set2"))(nb.cols)
# With barplot
RR_bar <- ggplot(agroforestry.analysed.imputed.prname.grouped, aes(x = reorder(PrName, -meanRR), y = meanRR, fill = PrName)) +
geom_bar(stat="identity", color="black", position = position_dodge()) +
geom_errorbar(aes(ymin = meanRR - meanRR.se, ymax = meanRR + meanRR.se), width=.2, position = position_dodge(.9)) +
ggplot2::scale_color_manual(values = era.af.colors) +
scale_y_continuous(expand = expand_scale(mult = c(0, 0.1))) +
geom_text(data = agroforestry.analysed.nstudies,
aes(x= reorder(PrName, -meanRR), y = n_studies_label_pos_RR, label = n_studies_1 , angle = 60),
check_overlap = FALSE, size = 4) +
theme_lucid(plot.title.size = 25,
axis.text.size = 15,
legend.title.size = 20,
legend.text.size = 20,
axis.title.size = 20) +
theme(legend.position = "none", axis.text.x = element_text(angle = 60, hjust=1), plot.margin=unit(c(1,6,1,6),"cm"))
# Create a text
no_studies <- grid::grobTree(textGrob("Number of studies", hjust = -3.2, y = 0.96,
gp = gpar(col="black", fontsize = 13, fontface="italic")))
# Finished bar ggplot bar plot
RR_bar + labs(title="Response ratio for agroforestry practices across all outcomes",
y = "Response Ratio (RR)",
x = "ERA agroforestry practice (PrName)") +
annotation_custom(no_studies)
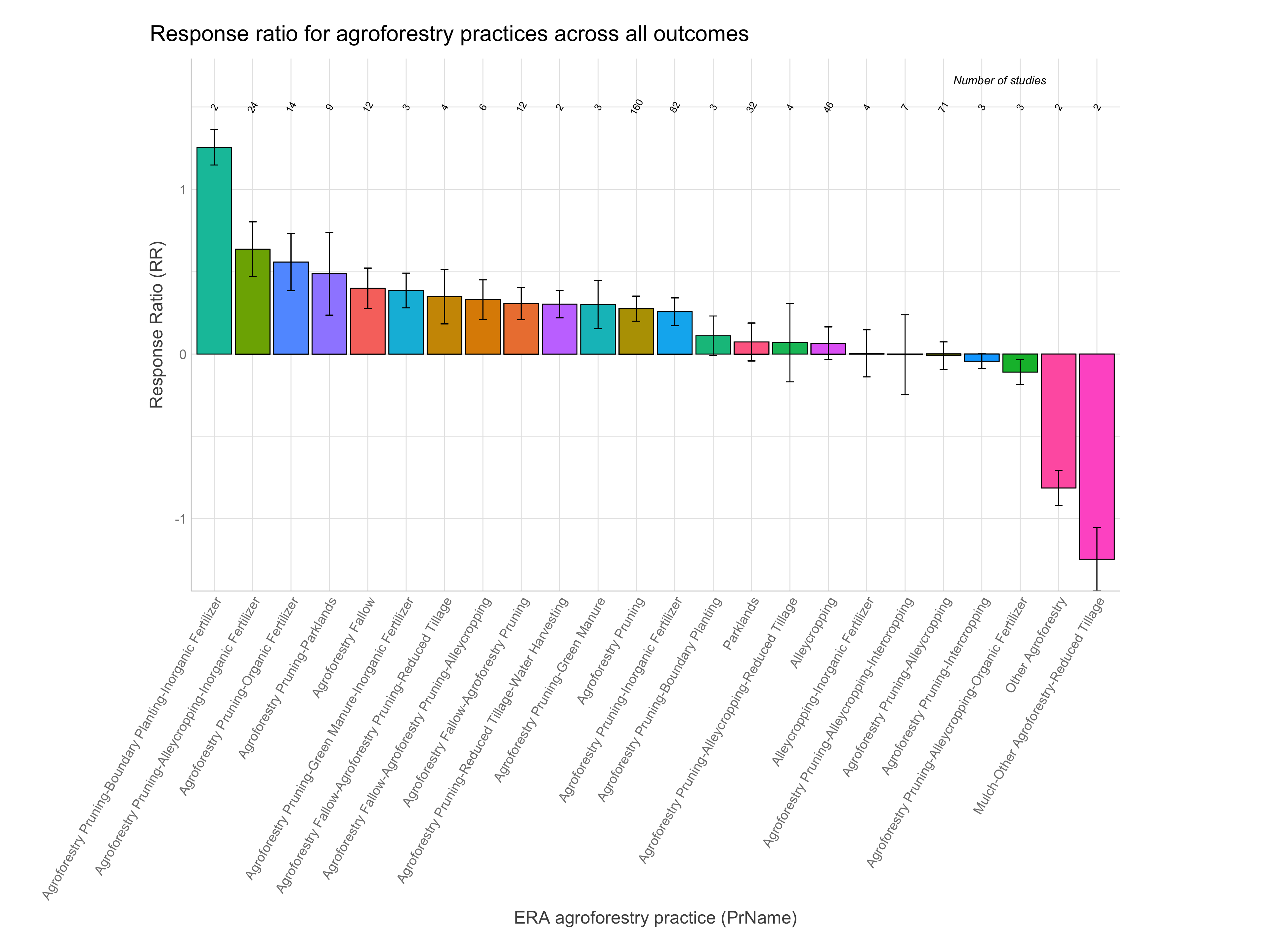
Figure 18: Proportion_MeanC/MeanT
Show code
mean(agroforestry.analysed.imputed.prname.grouped$meanRR.pc.jen)
[1] 34.06284Variations in RR.pc.jen for agroforestry practices
Plotting RR percentage with Jensen corrections for all agroforestry practices plot in bar plot
Show code
# With barplot
meanRR.pc.jen_bar <- ggplot(agroforestry.analysed.imputed.prname.grouped, aes(x = reorder(PrName, -meanRR.pc.jen), y = meanRR.pc.jen, fill = PrName)) +
geom_bar(stat="identity", color="black", position = position_dodge()) +
geom_errorbar(aes(ymin = meanRR.pc.jen.low, ymax = meanRR.pc.jen.high), width=.2, position = position_dodge(.9)) +
ggplot2::scale_color_manual(values = era.af.colors) +
scale_y_continuous(expand = expand_scale(mult = c(0, 0.1))) +
geom_text(data = agroforestry.analysed.nstudies,
aes(x= reorder(PrName, -meanRR), y = n_studies_label_pos_RR.pc.jen, label = n_studies_1, angle = 60),
check_overlap = FALSE, size = 4) +
theme_lucid(plot.title.size = 25,
axis.text.size = 15,
legend.title.size = 20,
legend.text.size = 20,
axis.title.size = 20) +
theme(legend.position = "none", axis.text.x = element_text(angle = 60, hjust=1), plot.margin=unit(c(1,6,1,6),"cm"))
# Create a text
no_studies <- grid::grobTree(textGrob("Number of studies", hjust = -3.2, y = 0.96,
gp = gpar(col="black", fontsize = 13, fontface="italic")))
# Finished bar ggplot bar plot
meanRR.pc.jen_bar + labs(title="RR.pc.jen for ERA agroforestry practices across all outcomes",
y = "% response ratios (RR) corrected for Jensen's inequality",
x = "ERA agroforestry practices (PrName)") +
annotation_custom(no_studies)
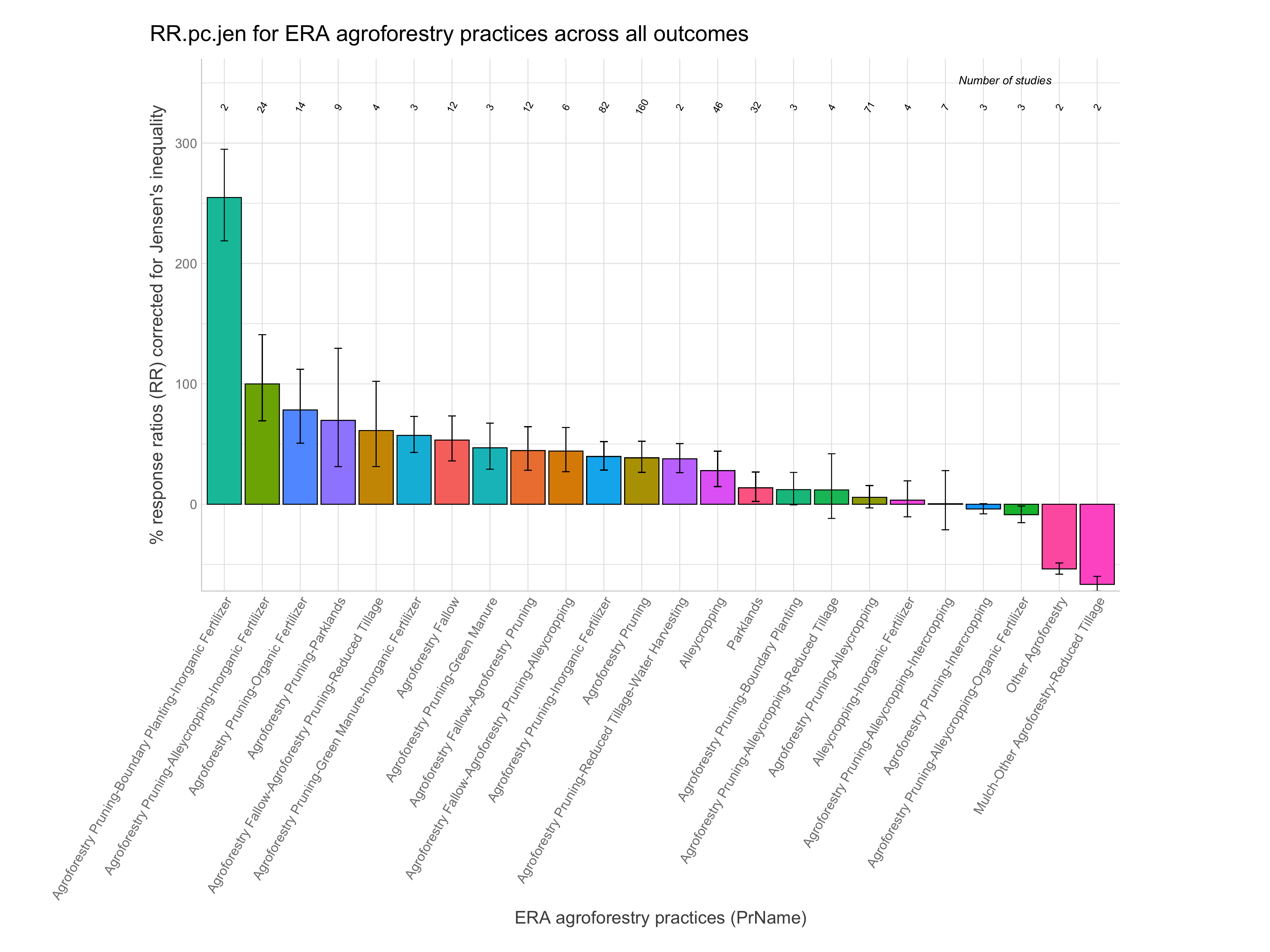
Figure 19: Proportion_MeanC/MeanT
Show code
# With barplot
meanRR.pc.jen_bar_selected <- agroforestry.analysed.imputed.prname.grouped %>%
ungroup() %>%
dplyr::mutate_if(sapply(agroforestry.analysed.imputed.prname.grouped, is.character), as.factor) %>%
dplyr::filter(PrName == "Agroforestry Fallow" |
PrName == "Agroforestry Pruning"|
PrName == "Alleycropping"|
PrName == "Parklands") %>%
ggplot(aes(x = reorder(PrName, -meanRR.pc.jen), y = meanRR.pc.jen, fill = PrName), palette = "Set2") +
geom_bar(stat="identity", color="black", position = position_dodge()) +
geom_errorbar(aes(ymin = meanRR.pc.jen.low, ymax = meanRR.pc.jen.high), width=.2, position = position_dodge(.9)) +
ggplot2::scale_color_manual(values = era.af.colors) +
scale_y_continuous(expand = expand_scale(mult = c(0, 0.1))) +
scale_fill_brewer(palette = "Set2") +
# geom_text(data = agroforestry.analysed.nstudies,
# aes(x= reorder(PrName, -meanRR), y = n_studies_label_pos_RR.pc.jen, label = n_studies_1, angle = 60),
# check_overlap = FALSE, size = 4) +
theme_lucid(plot.title.size = 25,
axis.text.size = 15,
legend.title.size = 20,
legend.text.size = 20,
axis.title.size = 20) +
theme(legend.position = "none", axis.text.x = element_text(angle = 60, hjust=1), plot.margin=unit(c(1,6,1,6),"cm"))
# Create a text
no_studies <- grid::grobTree(textGrob("Number of studies", hjust = -3.2, y = 0.96,
gp = gpar(col="black", fontsize = 13, fontface="italic")))
# Finished bar ggplot bar plot
meanRR.pc.jen_bar_selected + labs(title="RR.pc.jen for ERA agroforestry practices across all outcomes",
y = "% response ratios (RR) corrected for Jensen's inequality",
x = "ERA agroforestry practices (PrName)") +
annotation_custom(no_studies)

Show code
meanRR.pc.jen_bar_selected <- agroforestry.analysed.imputed.prname.grouped %>%
ungroup() %>%
dplyr::mutate_if(sapply(agroforestry.analysed.imputed.prname.grouped, is.character), as.factor) %>%
dplyr::filter(PrName == "Agroforestry Fallow" |
PrName == "Agroforestry Pruning"|
PrName == "Alleycropping"|
PrName == "Parklands") %>%
group_by(PrName) %>%
summarise(across(everything(), mean)) %>%
arrange(desc(meanRR.pc.jen))
meanRR.pc.jen_bar_selected
# A tibble: 4 × 8
PrName meanRR meanRR.se meanRR.pc.jen meanRR.pc.jen.l…
<fct> <dbl> <dbl> <dbl> <dbl>
1 Agroforestry Fallow 0.399 0.123 53.3 36.0
2 Agroforestry Pruning 0.275 0.0756 38.5 26.5
3 Alleycropping 0.0654 0.100 27.9 14.7
4 Parklands 0.0734 0.115 13.6 2.28
# … with 3 more variables: meanRR.pc.jen.high <dbl>, n_obs <dbl>,
# n_studies <dbl>RR.pc.jen is grouped mean for each practice across all outcome.
Variations in RR for agroforestry practices accros Crop and Biomass Yield outcomes
It is not always meaningful to look across all outcomes. Lets look at only for outcomes of crop yield and biomass yield
We do this by selecting data from the ‘agroforestry.analyzed.imputed’ that only has Biomass and Crop Yield outcomes
Show code
agroforestry.analyzed.imputed.yield <- agroforestry.analyzed.imputed %>%
dplyr::filter(Out.SubInd == "Biomass Yield" | Out.SubInd == "Crop Yield") %>%
group_by(PrName) %>%
dplyr::summarise(meanRR = mean(RR),
meanRR.se = mean(RR.se),
meanRR.pc.jen = mean(impute_RR.pc.jen),
meanRR.pc.jen.low = mean(impute_RR.pc.jen.low),
meanRR.pc.jen.high = mean(impute_RR.pc.jen.high),
n_obs = Observations,
n_studies = Studies)
agroforestry.analysed.nstudies.yield <- agroforestry.analyzed.imputed.yield %>%
group_by(PrName, meanRR, meanRR.pc.jen) %>% # <- grouping by PrName anf meanRR and meanRR.pc.jen with the assuption that they are all unique
summarise(across(n_obs, list(sum)),
across(n_studies, list(sum))) %>% # <- summarising the number of observations for each combination
mutate(n_obs_label_pos_RR = (n() * 1.5)) %>% # <- making a new column used solely for positioning the n_obs labes on the following bar plots
mutate(n_obs_label_pos_RR.pc.jen = n() * 330) %>%
mutate(n_studies_label_pos_RR = (n() * 1.5)) %>% # <- making a new column used solely for positioning the n_obs labes on the following bar plots
mutate(n_studies_label_pos_RR.pc.jen = n() * 330)
Plotting RR for all agroforestry practices with Biomass and Crop Yield outcomes
Show code
# Defining a custom colours range
# nb.cols <- 25
# era.af.colors <- colorRampPalette(brewer.pal(8, "Set2"))(nb.cols)
# With barplot
RR_bar_yield <- ggplot(agroforestry.analyzed.imputed.yield,
aes(x = reorder(PrName, -meanRR), y = meanRR, fill = PrName)) +
geom_bar(stat="identity", color="black", position = position_dodge()) +
geom_errorbar(aes(ymin = meanRR - meanRR.se, ymax = meanRR + meanRR.se), width=.2, position = position_dodge(.9)) +
ggplot2::scale_color_manual(values = era.af.colors) +
scale_y_continuous(expand = expand_scale(mult = c(0, 0.1))) +
geom_text(data = agroforestry.analysed.nstudies.yield,
aes(x= reorder(PrName, -meanRR), y = n_studies_label_pos_RR, label = n_studies_1 , angle = 60),
check_overlap = FALSE, size = 4) +
theme_lucid(plot.title.size = 25,
axis.text.size = 15,
legend.title.size = 20,
legend.text.size = 20,
axis.title.size = 20) +
theme(legend.position = "none", axis.text.x = element_text(angle = 60, hjust=1), plot.margin=unit(c(1,6,1,6),"cm"))
# Create a text
no_studies <- grid::grobTree(textGrob("Number of studies", hjust = -3.2, y = 0.96,
gp = gpar(col="black", fontsize = 13, fontface="italic")))
# Finished bar ggplot bar plot
RR_bar_yield + labs(title="Response ratio for agroforestry practices across Crop and Biomass Yield",
y = "Response Ratio (RR)",
x = "ERA agroforestry practice (PrName)") +
annotation_custom(no_studies)
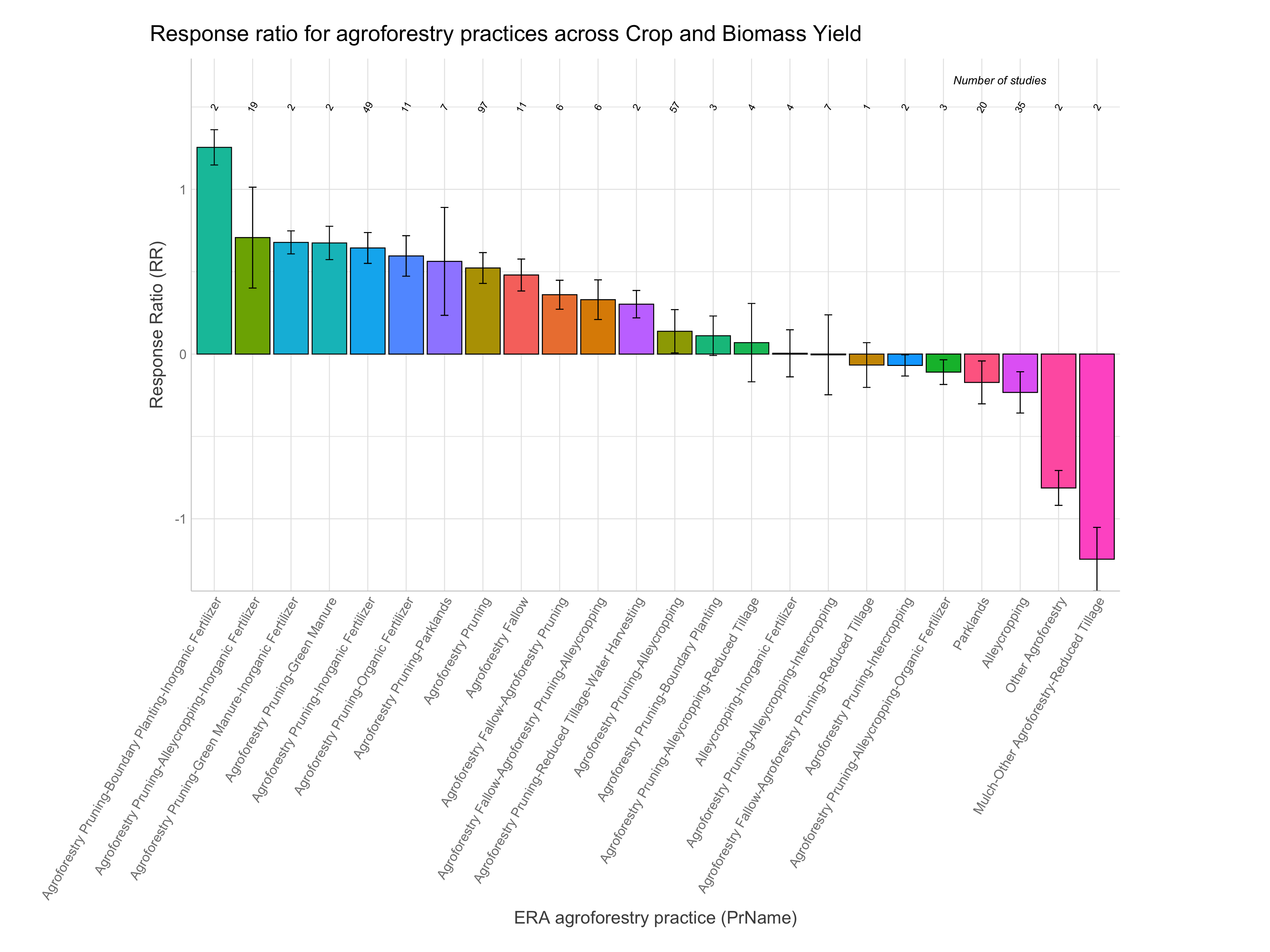
Figure 20: Proportion_MeanC/MeanT
Show code
# Warning: `expand_scale()` is deprecated; use `expansion()` instead.
Plotting RR.pc.jen for all agroforestry practices with Biomass and Crop Yield outcomes
Show code
# With barplot
RR.pc.jen_bar_yield <- ggplot(agroforestry.analyzed.imputed.yield,
aes(x = reorder(PrName, -meanRR.pc.jen), y = meanRR.pc.jen, fill = PrName)) +
geom_bar(stat="identity", color="black", position = position_dodge()) +
geom_errorbar(aes(ymin = meanRR.pc.jen.low, ymax = meanRR.pc.jen.high), width=.2, position = position_dodge(.9)) +
ggplot2::scale_color_manual(values = era.af.colors) +
scale_y_continuous(expand = expand_scale(mult = c(0, 0.1))) +
geom_text(data = agroforestry.analysed.nstudies.yield,
aes(x= reorder(PrName, -meanRR), y = n_studies_label_pos_RR.pc.jen, label = n_studies_1, angle = 60),
check_overlap = FALSE, size = 4) +
theme_lucid(plot.title.size = 25,
axis.text.size = 15,
legend.title.size = 20,
legend.text.size = 20,
axis.title.size = 20) +
theme(legend.position = "none", axis.text.x = element_text(angle = 60, hjust=1), plot.margin=unit(c(1,6,1,6),"cm"))
# Create a text
no_studies <- grid::grobTree(textGrob("Number of studies", hjust = -3.2, y = 0.96,
gp = gpar(col="black", fontsize = 13, fontface="italic")))
# Finished bar ggplot bar plot
RR.pc.jen_bar_yield + labs(title="RR.pc.jen for ERA agroforestry practices across Crop and Biomass Yield",
y = "% response ratios (RR) corrected for Jensen's inequality",
x = "ERA agroforestry practices (PrName)") +
annotation_custom(no_studies)
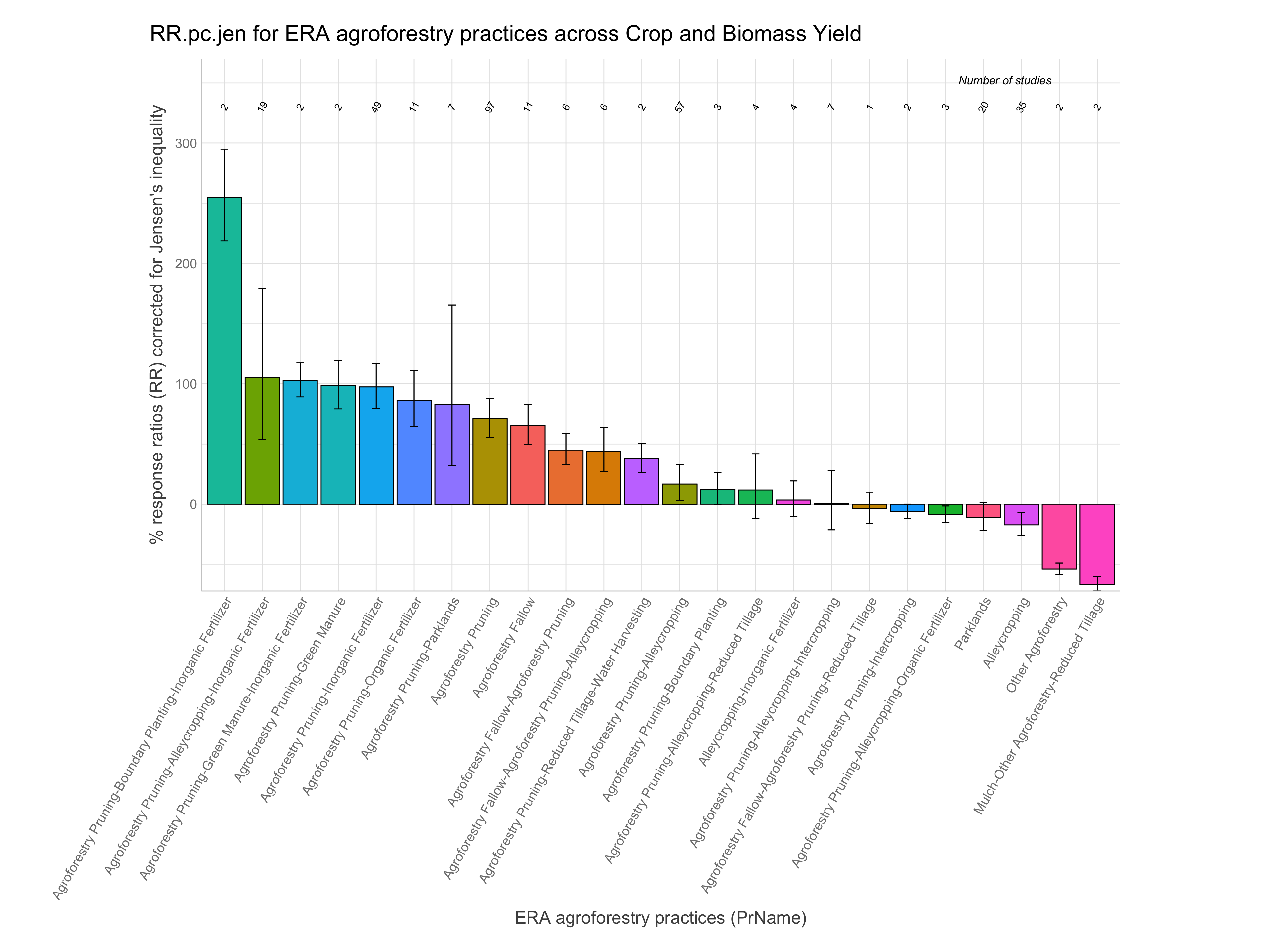
Figure 21: Proportion_MeanC/MeanT
Show code
ggplot(agroforestry.analyzed.imputed.yield, aes(x = meanRR.pc.jen, y = PrName)) +
geom_density_ridges()
Show code
agroforestry.analyzed.imputed.yield
# A tibble: 42 × 8
# Groups: PrName [24]
PrName meanRR meanRR.se meanRR.pc.jen meanRR.pc.jen.l…
<chr> <dbl> <dbl> <dbl> <dbl>
1 Agroforestry Fall… 0.480 0.0968 65.1 49.6
2 Agroforestry Fall… 0.480 0.0968 65.1 49.6
3 Agroforestry Fall… 0.360 0.0878 45.0 32.8
4 Agroforestry Fall… 0.360 0.0878 45.0 32.8
5 Agroforestry Fall… 0.330 0.120 44.1 27.0
6 Agroforestry Fall… 0.330 0.120 44.1 27.0
7 Agroforestry Fall… -0.0666 0.136 -3.82 -16.0
8 Agroforestry Prun… 0.522 0.0935 70.8 55.7
9 Agroforestry Prun… 0.522 0.0935 70.8 55.7
10 Agroforestry Prun… 0.138 0.131 16.8 2.78
# … with 32 more rows, and 3 more variables:
# meanRR.pc.jen.high <dbl>, n_obs <int>, n_studies <int>Variations in RR for agroforestry practices accros other outcomes
Soil Nitrogen and Total Soil Nitrogen
We do this by selecting data from the ‘agroforestry.analyzed.imputed’ that only has Soil Nitrogen and Total Soil Nitrogen outcomes
Show code
agroforestry.analyzed.imputed.nitrogen <- agroforestry.analyzed.imputed %>%
dplyr::filter(Out.SubInd == "Soil Nitrogen" | Out.SubInd == "Soil Nitrogen") %>%
group_by(PrName) %>%
dplyr::summarise(meanRR = mean(RR),
meanRR.se = mean(RR.se),
meanRR.pc.jen = mean(impute_RR.pc.jen),
meanRR.pc.jen.low = mean(impute_RR.pc.jen.low),
meanRR.pc.jen.high = mean(impute_RR.pc.jen.high),
n_obs = Observations,
n_studies = Studies)
agroforestry.analysed.nstudies.nitrogen <- agroforestry.analyzed.imputed.nitrogen %>%
group_by(PrName, meanRR, meanRR.pc.jen) %>% # <- grouping by PrName anf meanRR and meanRR.pc.jen with the assuption that they are all unique
summarise(across(n_obs, list(sum)),
across(n_studies, list(sum))) %>% # <- summarising the number of observations for each combination
mutate(n_obs_label_pos_RR = (n() * 1.5)) %>% # <- making a new column used solely for positioning the n_obs labes on the following bar plots
mutate(n_obs_label_pos_RR.pc.jen = n() * 330) %>%
mutate(n_studies_label_pos_RR = (n() * 1.5)) %>% # <- making a new column used solely for positioning the n_obs labes on the following bar plots
mutate(n_studies_label_pos_RR.pc.jen = n() * 330)
Plotting RR for all agroforestry practices with Soil Nitrogen and Total Soil Nitrogen outcomes
Show code
# Defining a custom colours range
# nb.cols <- 25
# era.af.colors <- colorRampPalette(brewer.pal(8, "Set2"))(nb.cols)
# With barplot
RR_bar_nitrogen <- ggplot(agroforestry.analyzed.imputed.nitrogen,
aes(x = reorder(PrName, -meanRR), y = meanRR, fill = PrName)) +
geom_bar(stat="identity", color="black", position = position_dodge()) +
geom_errorbar(aes(ymin = meanRR - meanRR.se, ymax = meanRR + meanRR.se), width=.2, position = position_dodge(.9)) +
ggplot2::scale_color_manual(values = era.af.colors) +
scale_y_continuous(expand = expand_scale(mult = c(0, 0.1))) +
geom_text(data = agroforestry.analysed.nstudies.nitrogen,
aes(x= reorder(PrName, -meanRR), y = n_studies_label_pos_RR, label = n_studies_1 , angle = 60),
check_overlap = FALSE, size = 4) +
theme_lucid(plot.title.size = 25,
axis.text.size = 15,
legend.title.size = 20,
legend.text.size = 20,
axis.title.size = 20) +
theme(legend.position = "none", axis.text.x = element_text(angle = 60, hjust=1), plot.margin=unit(c(1,6,1,6),"cm"))
# Create a text
no_studies <- grid::grobTree(textGrob("Number of studies", hjust = -3.2, y = 0.96,
gp = gpar(col="black", fontsize = 13, fontface="italic")))
# Finished bar ggplot bar plot
RR_bar_nitrogen + labs(title="Response ratio for agroforestry practices across Soil Nitrogen and Total Soil Nitrogen",
y = "Response Ratio (RR)",
x = "ERA agroforestry practice (PrName)") +
annotation_custom(no_studies)
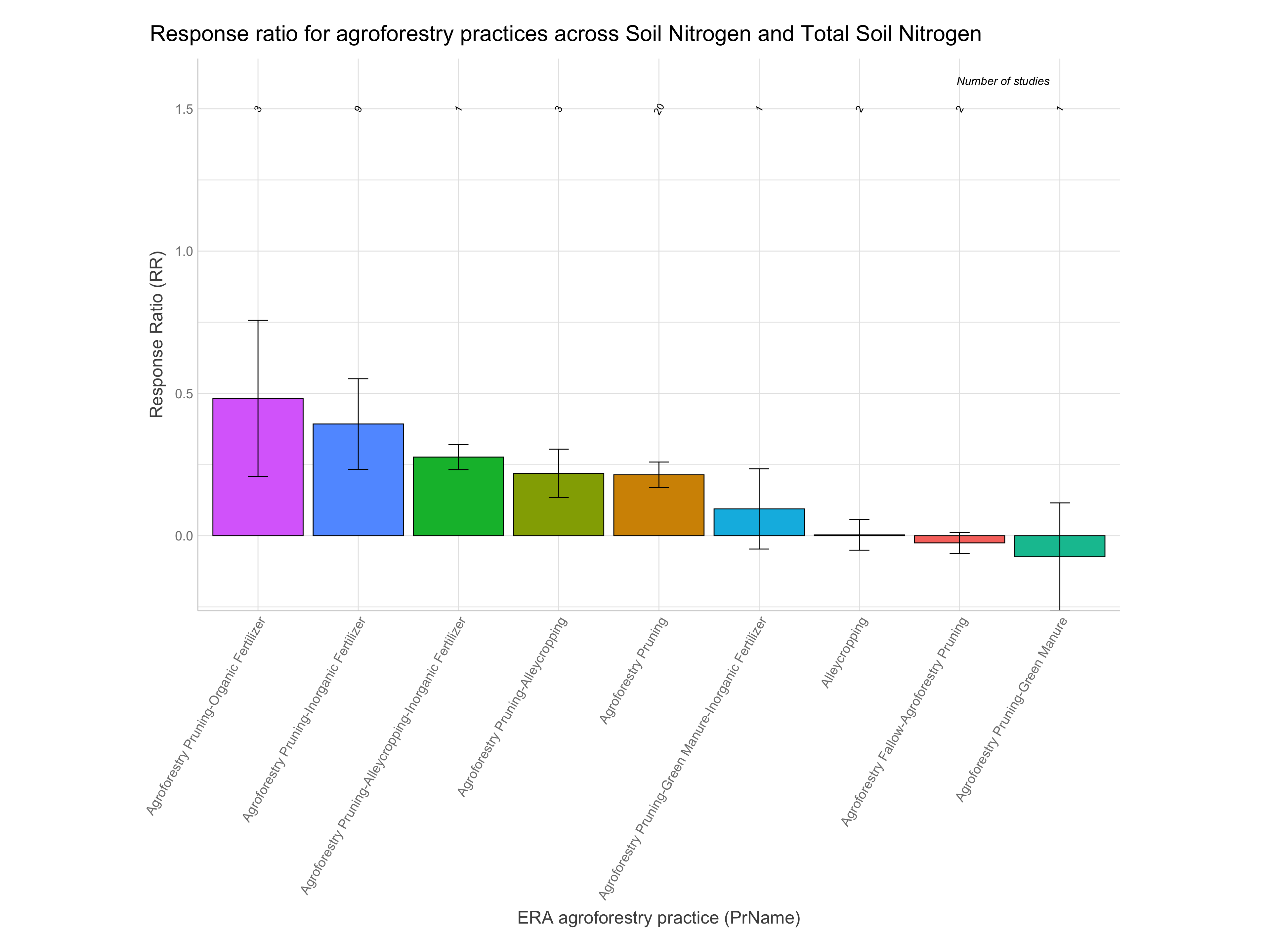
Figure 22: Proportion_MeanC/MeanT
Show code
# Warning: `expand_scale()` is deprecated; use `expansion()` instead.
Plotting RR.pc.jen for all agroforestry practices with Soil Nitrogen and Total Soil Nitrogen outcomes
Show code
# With barplot
RR.pc.jen_bar_nitrogen <- ggplot(agroforestry.analyzed.imputed.nitrogen,
aes(x = reorder(PrName, -meanRR.pc.jen), y = meanRR.pc.jen, fill = PrName)) +
geom_bar(stat="identity", color="black", position = position_dodge()) +
geom_errorbar(aes(ymin = meanRR.pc.jen.low, ymax = meanRR.pc.jen.high), width=.2, position = position_dodge(.9)) +
ggplot2::scale_color_manual(values = era.af.colors) +
scale_y_continuous(expand = expand_scale(mult = c(0, 0.1))) +
geom_text(data = agroforestry.analysed.nstudies.nitrogen,
aes(x= reorder(PrName, -meanRR), y = n_studies_label_pos_RR.pc.jen, label = n_studies_1, angle = 60),
check_overlap = FALSE, size = 4) +
theme_lucid(plot.title.size = 25,
axis.text.size = 15,
legend.title.size = 20,
legend.text.size = 20,
axis.title.size = 20) +
theme(legend.position = "none", axis.text.x = element_text(angle = 60, hjust=1), plot.margin=unit(c(1,6,1,6),"cm"))
# Create a text
no_studies <- grid::grobTree(textGrob("Number of studies", hjust = -3.2, y = 0.96,
gp = gpar(col="black", fontsize = 13, fontface="italic")))
# Finished bar ggplot bar plot
RR.pc.jen_bar_nitrogen + labs(title="RR.pc.jen for ERA agroforestry practices across Soil Nitrogen and Total Soil Nitroge",
y = "% response ratios (RR) corrected for Jensen's inequality",
x = "ERA agroforestry practices (PrName)") +
annotation_custom(no_studies)
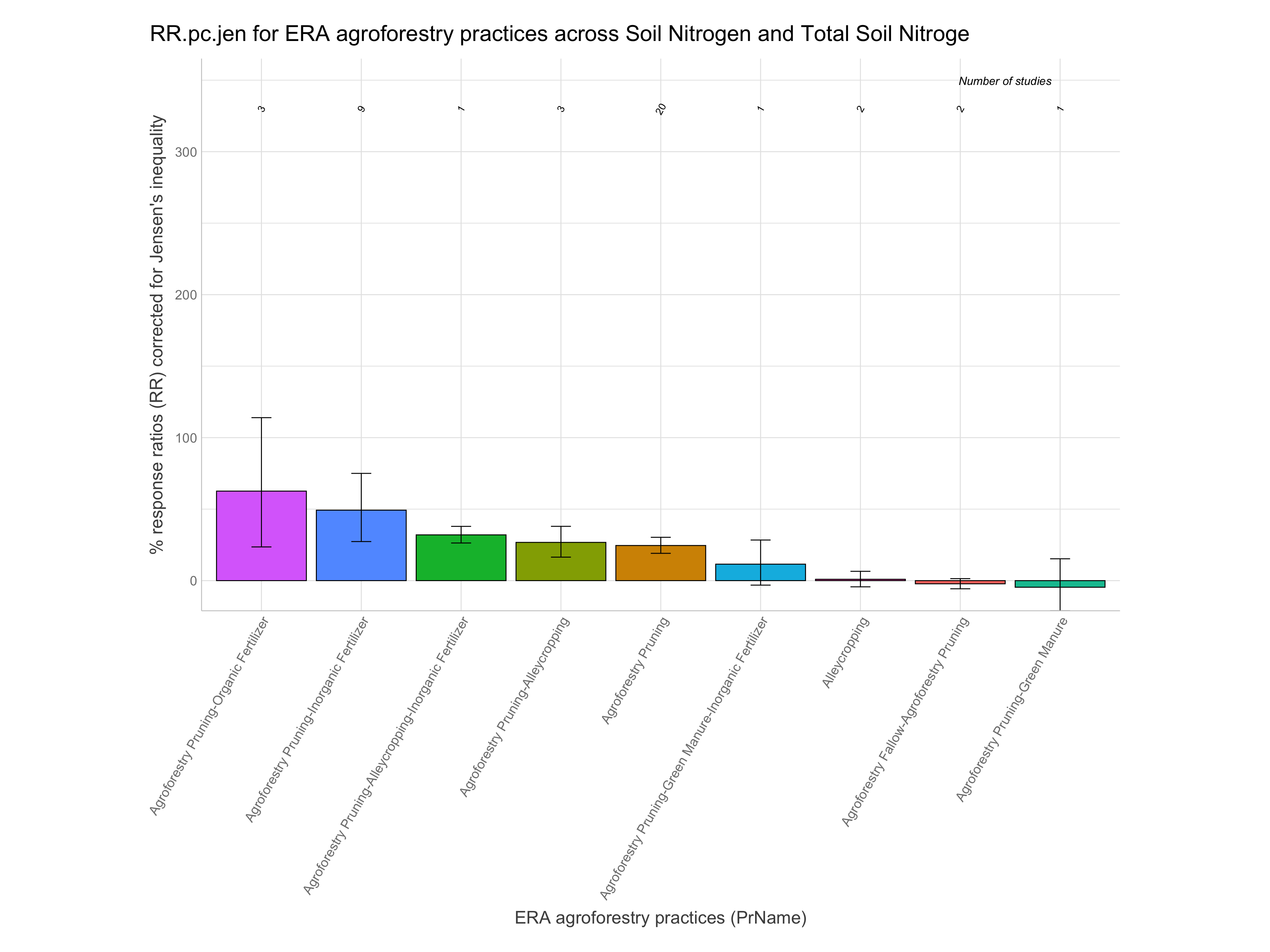
Figure 23: Proportion_MeanC/MeanT
Water Use Efficiency
We do this by selecting data from the ‘agroforestry.analyzed.imputed’ that only has Water Use Efficiency outcomes
Show code
agroforestry.analyzed.imputed.wue <- agroforestry.analyzed.imputed %>%
dplyr::filter(Out.SubInd == "Water Use Efficiency") %>%
group_by(PrName) %>%
dplyr::summarise(meanRR = mean(RR),
meanRR.se = mean(RR.se),
meanRR.pc.jen = mean(impute_RR.pc.jen),
meanRR.pc.jen.low = mean(impute_RR.pc.jen.low),
meanRR.pc.jen.high = mean(impute_RR.pc.jen.high),
n_obs = Observations,
n_studies = Studies)
agroforestry.analysed.nstudies.wue <- agroforestry.analyzed.imputed.wue %>%
group_by(PrName, meanRR, meanRR.pc.jen) %>% # <- grouping by PrName anf meanRR and meanRR.pc.jen with the assuption that they are all unique
summarise(across(n_obs, list(sum)),
across(n_studies, list(sum))) %>% # <- summarising the number of observations for each combination
mutate(n_obs_label_pos_RR = (n() * 1.5)) %>% # <- making a new column used solely for positioning the n_obs labes on the following bar plots
mutate(n_obs_label_pos_RR.pc.jen = n() * 330) %>%
mutate(n_studies_label_pos_RR = (n() * 1.5)) %>% # <- making a new column used solely for positioning the n_obs labes on the following bar plots
mutate(n_studies_label_pos_RR.pc.jen = n() * 330)
Plotting RR for all agroforestry practices with Water Use Effeciency outcomes
Show code
# Defining a custom colours range
# nb.cols <- 25
# era.af.colors <- colorRampPalette(brewer.pal(8, "Set2"))(nb.cols)
# With barplot
RR_bar_wue <- ggplot(agroforestry.analyzed.imputed.wue,
aes(x = reorder(PrName, -meanRR), y = meanRR, fill = PrName)) +
geom_bar(stat="identity", color="black", position = position_dodge()) +
geom_errorbar(aes(ymin = meanRR - meanRR.se, ymax = meanRR + meanRR.se), width=.2, position = position_dodge(.9)) +
ggplot2::scale_color_manual(values = era.af.colors) +
scale_y_continuous(expand = expand_scale(mult = c(0, 0.1))) +
geom_text(data = agroforestry.analysed.nstudies.wue,
aes(x= reorder(PrName, -meanRR), y = n_studies_label_pos_RR, label = n_studies_1 , angle = 60),
check_overlap = FALSE, size = 4) +
theme_lucid(plot.title.size = 25,
axis.text.size = 15,
legend.title.size = 20,
legend.text.size = 20,
axis.title.size = 20) +
theme(legend.position = "none", axis.text.x = element_text(angle = 60, hjust=1), plot.margin=unit(c(1,6,1,6),"cm"))
# Create a text
no_studies <- grid::grobTree(textGrob("Number of studies", hjust = -3.2, y = 0.96,
gp = gpar(col="black", fontsize = 13, fontface="italic")))
# Finished bar ggplot bar plot
RR_bar_wue + labs(title="Response ratio for agroforestry practices across Water Use Effeciency",
y = "Response Ratio (RR)",
x = "ERA agroforestry practice (PrName)") +
annotation_custom(no_studies)
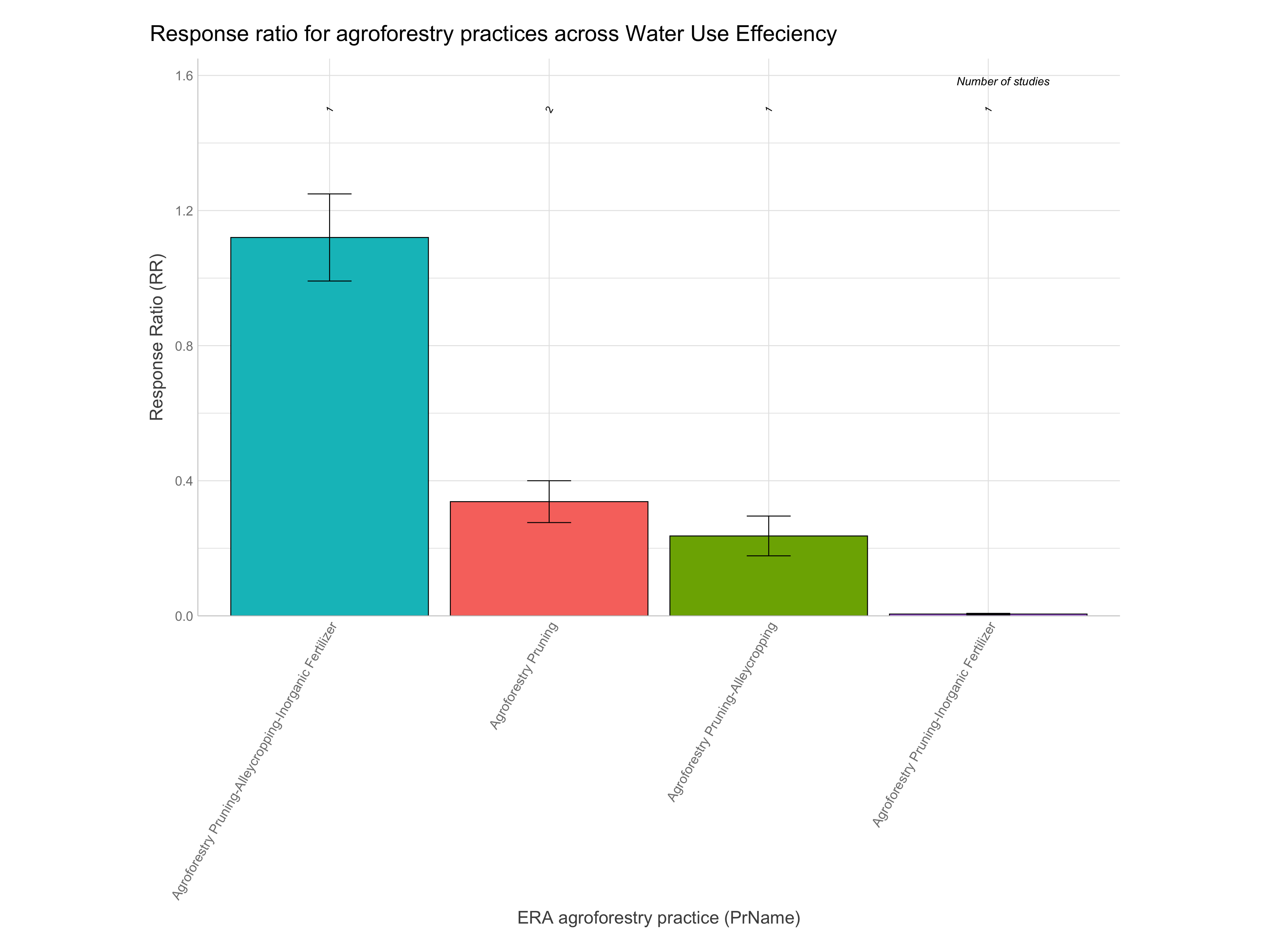
Figure 24: Proportion_MeanC/MeanT
Show code
# Warning: `expand_scale()` is deprecated; use `expansion()` instead.
Plotting RR.pc.jen for all agroforestry practices with Water Use Effeciency outcomes
Show code
# With barplot
RR.pc.jen_bar_wue <- ggplot(agroforestry.analyzed.imputed.wue,
aes(x = reorder(PrName, -meanRR.pc.jen), y = meanRR.pc.jen, fill = PrName)) +
geom_bar(stat="identity", color="black", position = position_dodge()) +
geom_errorbar(aes(ymin = meanRR.pc.jen.low, ymax = meanRR.pc.jen.high), width=.2, position = position_dodge(.9)) +
ggplot2::scale_color_manual(values = era.af.colors) +
scale_y_continuous(expand = expand_scale(mult = c(0, 0.1))) +
geom_text(data = agroforestry.analysed.nstudies.wue,
aes(x= reorder(PrName, -meanRR), y = n_studies_label_pos_RR.pc.jen, label = n_studies_1, angle = 60),
check_overlap = FALSE, size = 4) +
theme_lucid(plot.title.size = 25,
axis.text.size = 15,
legend.title.size = 20,
legend.text.size = 20,
axis.title.size = 20) +
theme(legend.position = "none", axis.text.x = element_text(angle = 60, hjust=1), plot.margin=unit(c(1,6,1,6),"cm"))
# Create a text
no_studies <- grid::grobTree(textGrob("Number of studies", hjust = -3.2, y = 0.96,
gp = gpar(col="black", fontsize = 13, fontface="italic")))
# Finished bar ggplot bar plot
RR.pc.jen_bar_wue + labs(title="RR.pc.jen for ERA agroforestry practices across Water Use Effeciency",
y = "% response ratios (RR) corrected for Jensen's inequality",
x = "ERA agroforestry practices (PrName)") +
annotation_custom(no_studies)
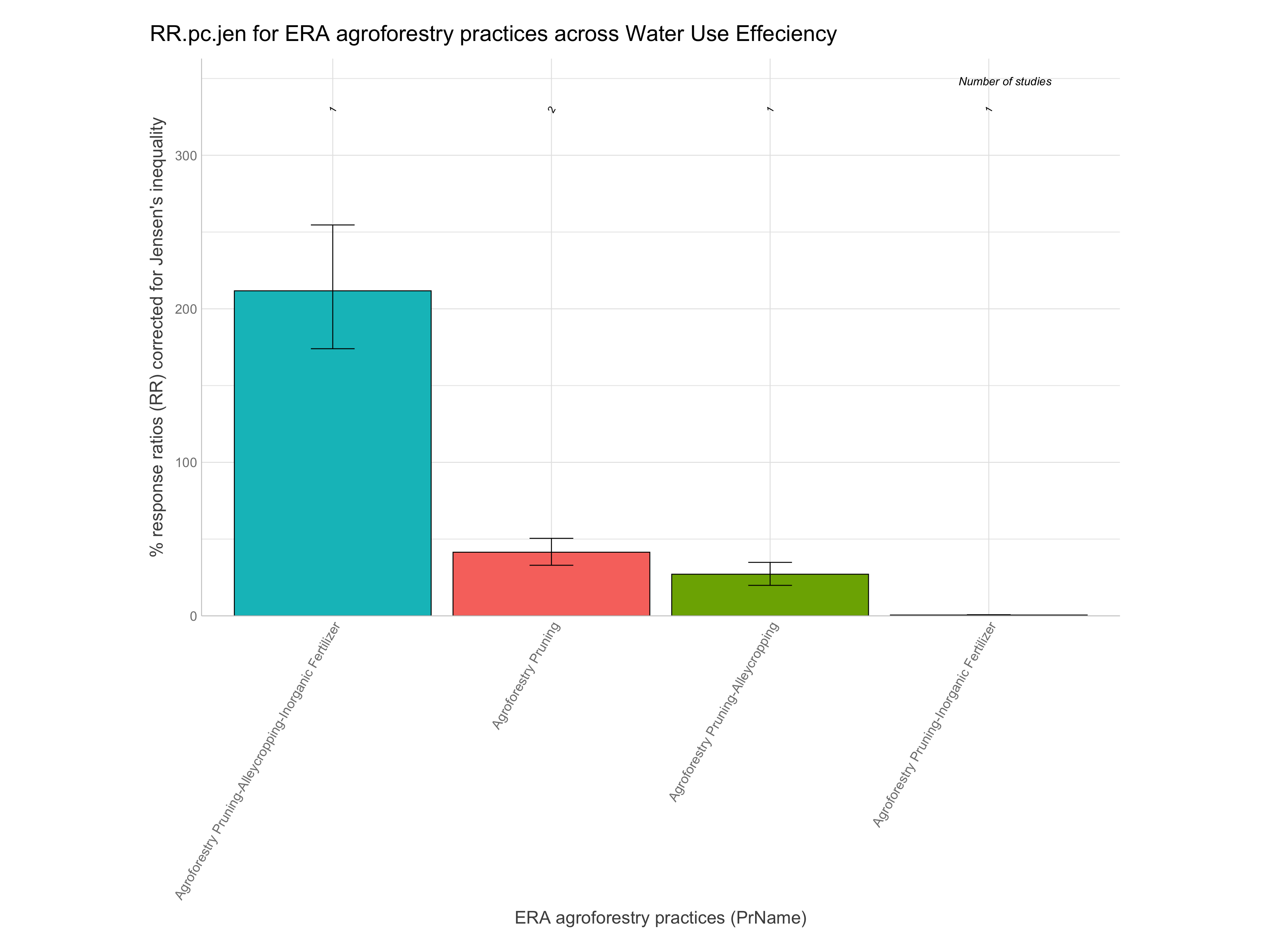
Figure 25: Proportion_MeanC/MeanT
Soil Organic Carbon and Soil Organic Matter and Soil Carbon Stocks
We do this by selecting data from the ‘agroforestry.analyzed.imputed’ that only has Soil Organic Carbon and Soil Organic Matter and Soil Carbon Stocks outcomes
Show code
agroforestry.analyzed.imputed.org <- agroforestry.analyzed.imputed %>%
dplyr::filter(Out.SubInd == "Soil Organic Carbon" |
Out.SubInd == "Soil Organic Matter" |
Out.SubInd == "Soil Carbon Stocks") %>%
group_by(PrName) %>%
dplyr::summarise(meanRR = mean(RR),
meanRR.se = mean(RR.se),
meanRR.pc.jen = mean(impute_RR.pc.jen),
meanRR.pc.jen.low = mean(impute_RR.pc.jen.low),
meanRR.pc.jen.high = mean(impute_RR.pc.jen.high),
n_obs = Observations,
n_studies = Studies)
agroforestry.analysed.nstudies.org <- agroforestry.analyzed.imputed.org %>%
group_by(PrName, meanRR, meanRR.pc.jen) %>% # <- grouping by PrName anf meanRR and meanRR.pc.jen with the assuption that they are all unique
summarise(across(n_obs, list(sum)),
across(n_studies, list(sum))) %>% # <- summarising the number of observations for each combination
mutate(n_obs_label_pos_RR = (n() * 1.5)) %>% # <- making a new column used solely for positioning the n_obs labes on the following bar plots
mutate(n_obs_label_pos_RR.pc.jen = n() * 330) %>%
mutate(n_studies_label_pos_RR = (n() * 1.5)) %>% # <- making a new column used solely for positioning the n_obs labes on the following bar plots
mutate(n_studies_label_pos_RR.pc.jen = n() * 330)
Plotting RR for all agroforestry practices with Soil Organic Carbon and Soil Organic Matter and Soil Carbon Stocks outcomes
Show code
# Defining a custom colours range
# nb.cols <- 25
# era.af.colors <- colorRampPalette(brewer.pal(8, "Set2"))(nb.cols)
# With barplot
RR_bar_org <- ggplot(agroforestry.analyzed.imputed.org,
aes(x = reorder(PrName, -meanRR), y = meanRR, fill = PrName)) +
geom_bar(stat="identity", color="black", position = position_dodge()) +
geom_errorbar(aes(ymin = meanRR - meanRR.se, ymax = meanRR + meanRR.se), width=.2, position = position_dodge(.9)) +
ggplot2::scale_color_manual(values = era.af.colors) +
scale_y_continuous(expand = expand_scale(mult = c(0, 0.1))) +
geom_text(data = agroforestry.analysed.nstudies.org,
aes(x= reorder(PrName, -meanRR), y = n_studies_label_pos_RR, label = n_studies_1 , angle = 60),
check_overlap = FALSE, size = 4) +
theme_lucid(plot.title.size = 25,
axis.text.size = 15,
legend.title.size = 20,
legend.text.size = 20,
axis.title.size = 20) +
theme(legend.position = "none", axis.text.x = element_text(angle = 60, hjust=1), plot.margin=unit(c(1,6,1,6),"cm"))
# Create a text
no_studies <- grid::grobTree(textGrob("Number of studies", hjust = -3.2, y = 0.96,
gp = gpar(col="black", fontsize = 13, fontface="italic")))
# Finished bar ggplot bar plot
RR_bar_org + labs(title="Response ratio for agroforestry practices across Soil Organic Carbon and Soil Organic Matter and Soil Carbon Stocks",
y = "Response Ratio (RR)",
x = "ERA agroforestry practice (PrName)") +
annotation_custom(no_studies)
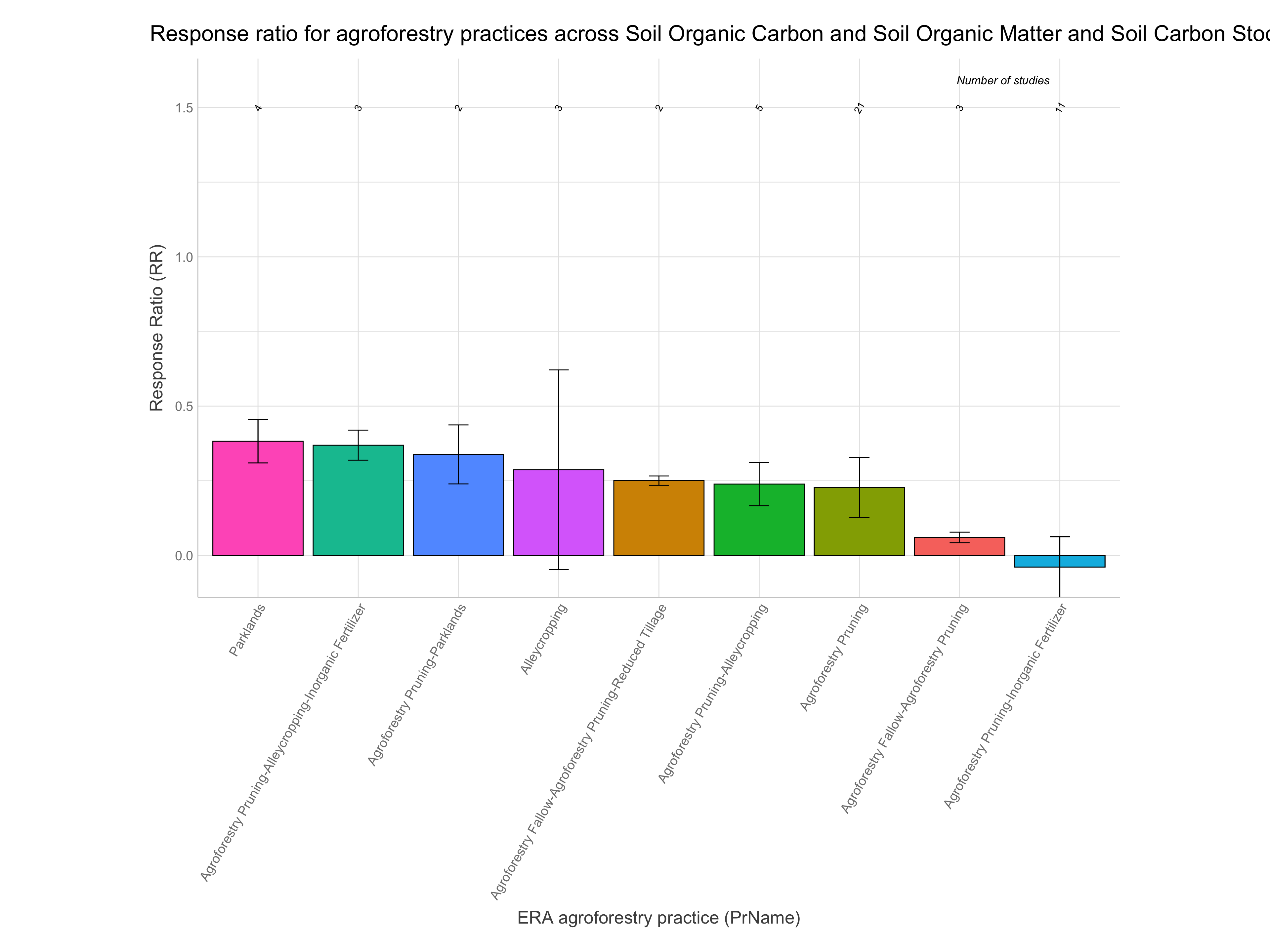
Figure 26: Proportion_MeanC/MeanT
Show code
# Warning: `expand_scale()` is deprecated; use `expansion()` instead.
Plotting RR.pc.jen for all agroforestry practices with Soil Organic Carbon and Soil Organic Matter and Soil Carbon Stocks outcomes
Show code
# With barplot
RR.pc.jen_bar_org <- ggplot(agroforestry.analyzed.imputed.org,
aes(x = reorder(PrName, -meanRR.pc.jen), y = meanRR.pc.jen, fill = PrName)) +
geom_bar(stat="identity", color="black", position = position_dodge()) +
geom_errorbar(aes(ymin = meanRR.pc.jen.low, ymax = meanRR.pc.jen.high), width=.2, position = position_dodge(.9)) +
ggplot2::scale_color_manual(values = era.af.colors) +
scale_y_continuous(expand = expand_scale(mult = c(0, 0.1))) +
geom_text(data = agroforestry.analysed.nstudies.org,
aes(x= reorder(PrName, -meanRR), y = n_studies_label_pos_RR.pc.jen, label = n_studies_1, angle = 60),
check_overlap = FALSE, size = 4) +
theme_lucid(plot.title.size = 25,
axis.text.size = 15,
legend.title.size = 20,
legend.text.size = 20,
axis.title.size = 20) +
theme(legend.position = "none", axis.text.x = element_text(angle = 60, hjust=1), plot.margin=unit(c(1,6,1,6),"cm"))
# Create a text
no_studies <- grid::grobTree(textGrob("Number of studies", hjust = -3.2, y = 0.96,
gp = gpar(col="black", fontsize = 13, fontface="italic")))
# Finished bar ggplot bar plot
RR.pc.jen_bar_org + labs(title="RR.pc.jen for ERA agroforestry practices across Soil Organic Carbon and Soil Organic Matter and Soil Carbon Stocks",
y = "% response ratios (RR) corrected for Jensen's inequality",
x = "ERA agroforestry practices (PrName)") +
annotation_custom(no_studies)
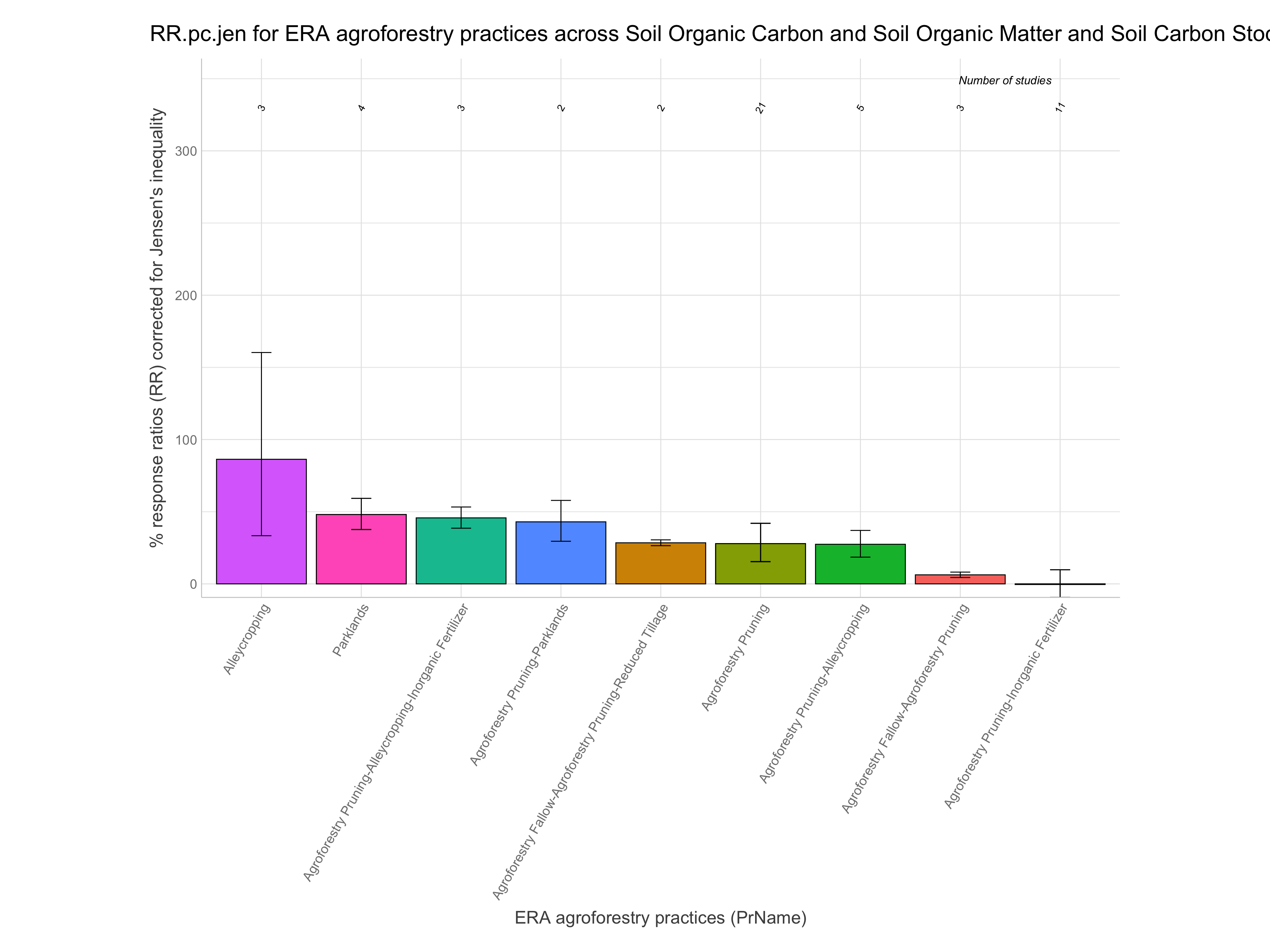
Figure 27: Proportion_MeanC/MeanT